| |
ID |
Date |
Author |
Subject |
|
|
17
|
Thu Nov 7 16:55:00 2019 |
Cade Sbrocco | Archiving Old Genetis Manual | |
|
|
11
|
Wed Apr 3 16:38:07 2019 |
Julie Rolla | AraSim update | General update
The following were the fixes we made to make AraSim run:
(1) As of March, the loop breaks at AraSim. We have found that our program XFintoARA.py does not properly format the output in the form that AraSim is looking for (as an input). Jorge gave us a sample AraSim input file to help us edit XFintoARA.py so that it formats the file correctly. Note that we found white space is also must match in order for the format to be correct.
Now that we have fixed this, AraSim now runs. Attached is the following sample Jorge gave us of the proper AraSim inputs. If needed, it's available to refer back to.
(2) After correcting the AraSim input file formats (outputs of XFintoARA.py), we ran it. It failed to finish running after hours of run time. We found that we were using default input parameters for AraSim -- which wasn't triggering in a timely fashion. Once we edited the parameters, it ran successfully. Note that this was not run within the loop. Confirmation of the full loop running has not been completed.
The parameters of interest need to be edited in setup.txt. To see which file in AraSim I am referring to, please see the .txt file attached. Note that we will be editing the bash script to allow the user to change these input parameters there. The following red lines are what need to be set:
EXPONENT=21
NNU=100 // number of neutrino events
NNU_PASSED=10 // number of neutrino events that are allowed to pass the trigger
ONLY_PASSED_EVENTS=1 // 0 (default): AraSim throws NNU events whether or not they pass; 1: AraSim throws events until the number of events that pass the trigger is equal to NNU_PASSED (WARNING: may cause long run times if reasonable values are not chosen)
NOISE_WAVEFORM_GENERATE_MODE=0 // generate new noise waveforms for each events
NOISE_EVENTS=16 // number of pure noise waveforms
TRIG_ANALYSIS_MODE=0 // 0 = signal + noise, 1 = signal only, 2 = noise only
DETECTOR=1 // ARA stations 1 to 7
NOFZ=1
core_x=10000
core_y=10000
EXPONENT gives the energy of the neutrinos. Note that higher energies will have more pass, as the energies don't dissipate as rapidly. For now, we are running at this energy. Eventally, we will want to use the full Kotera spectrum. Otherwsie, we would be finding the performance of an antenna at only one energy. Just to check the loop, we are going to use EXPONENT=21 to get events to trigger faster.
ONLY_PASSED_EVENTS decides which of the two options to use to determine when AraSim is finished running: (1) Throws 'NNU' number of events at antennas and stops regardless of how many pass (in this case ONLY_PASSED_EVENTS=0) , or (2) throws as many events at the antenna as needed until 'NNU_PASSED' number triggers (in this case ONLY_PASSED_EVENTS=1). Ie if this number is 10, we throw as many neutrinos at the antenna as necessary until 10 events trigger. Note that this can result in a longer run time. Additionally, a higher EXPONENT will make them trigger faster.
TRIG_ANALYSIS_MODE is a setting that determines what is being thrown. Ie- 0=signal + noise (which is what we want, and is the 'real' case), 1=signal only (would make it run faster), 2=noise only (not of interest for us).
To-Do List
(1) Commit version of XFintoARA.py since recent formatting change.
(2) Figure out how to add in Kotera spectrum (note that we wont want to do this until the WHOLE loop is functional)
(3) Edit the bash script to recreat the setup.txt making the variables in red above an editable option in the top of the bash script. Ie our variables in red above will be at the top of the bash script. Later in the bash script we will need to have it create this .txt file with the user inputs for those variables. We can do this by either (1) creating a skeleton, or (2) creating another .sh script that is run in our XFLoop.sh -- which writes the first handfull of lines based on the user inputs for our variables in red above, and then pastes in the rest of what is unchanged in setup.txt.
(4) Run the whole loop: update- we tried this today and keep getting a Veff-0 for our first antenna along with the error
Warning in <TFile::Init>: no StreamerInfo found in /datapool/software/anita/lib/G__RootFftwWrapper_rdict.pcm therefore preventing schema evolution when reading this file.
Error in <TStreamerInfo::Build>: AraEventCalibrator, discarding: unsigned short* fAtriPeds, no [dimension]
others run as expected
(5) Work on the paper: Amy added a skeleton of sections to add for the GA. Date of completion should be in-line with APS.
(6) APS talk slides.
(7) Edit the manual and add:
- The changes that will occur when task (3) is done.
- A sample of how AraSim inputs -- or XFintoARA.py outputs -- should look.
- This stuff: http://radiorm.physics.ohio-state.edu/elog/GENETIS/8
- Max's AraSim plotting software (when it's done)
- Evelyn and Sophie's plotting software info (when it's done).
(8) Finish Max's AraSim plotting software.
(9) Finish Evenlyn and Sophie's plotting softare. |
|
|
197
|
Mon Feb 20 12:33:20 2023 |
Dennis H. Calderon | AraSim Simulations for Top 5 Antennas (12/2022) with Higher Stats | AraSim Simulations for Top 5 Antennas from 12/2022 Run with 3 million simulated events
Simulated using Arasim version currently used in GENETIS Loop
/fs/ess/PAS1960/BiconeEvolutionOSC/AraSim/
AraSim ROOT files stored in
/fs/ess/PAS1960/ROOT_Files_Higher_Stats/AraSim_Loop_122022/Feb_2023_Crazy_Sides/
Antennas simulated
- Generation: 13, Individual: 84
- Generation: 18, Individual: 89
- Generation: 19, Individual: 96
- Generation: 29, Individual: 87
- Generation: 9, Individual: 50
Results
| Antenna |
Total Events |
Triggered Events |
Weighted Events |
Effective Volume |
Effective Volume Error |
| 13_84 |
2995982 |
40251 |
16058 |
5.71 |
0.035 |
| 18_89 |
3010000 |
40224 |
15963 |
5.65 |
0.035 |
| 19_96 |
3010000 |
39989 |
16034 |
5.68 |
0.031 |
| 29_87 |
3010000 |
40056 |
16049 |
5.68 |
0.034 |
| 9_50 |
3010000 |
40042 |
16057 |
5.69 |
0.034 |
| ARA_Sym_Bicone_6in |
3010000 |
36627 |
14334 |
5.08 |
0.032 |
Conclusion
Antenna from Generation 13 Individual 84 performed the best with these stats |
|
|
240
|
Tue May 21 09:55:57 2024 |
Jacob Weiler | AraSim CSE Spring 2024 Work | # AraSim CSE Spring 2024 Work
## Goals
The main goal was to get a working multithreaded version of the AraSim codebase working. Doing this, the hope was to learn how to multithread the code and get it in a good place to hopefully also integrate GPU's at a later date.
## Where is it at currently?
A bulk of the work done was to functionalize the Connect_Interaction_Detector_V2 to allow for multithreading and to cleanup codebase.
### Going through code added/changed
Helper functions for multiple parts of code. They went through and split it up into multiple parts. Putting line numbers when needed
Part 1: Clearning Antenna Data
Part 2: Determine gain channel
Part 3: solve ray tracing (Not Done)
Part 4: Process Ray Tracing Solution (Not Done)
Part 5: Calculate Signal Factors (Not Done)
Part 6: Calculate Antenna Gain Factors (Not Done)
Part 7: Process Frequency Domain Signal
Part 8A: Process Neutrino Events. Lines 908 - 1208 (Not Done)
Part 8B: Process Arbitrary Events. Lines 1209 - 1475
Part 8C: Process Simple Pulser Simulation. Lines 1478 - 1747 (Not Done)
Part 8D: Process PVA Pulser Simulation. Lines 1751 - 2110 (Not Done)
Part 8E: Process Calpulser Event. Lines 2113 - 2445
Part 9A: Process Noise. Lines 2593 - 2955
Part 9B: Process Trigger and Mimic Waveforms. Lines 2994 - 3624
## What still needs to be done?
- Multithreading still isn't working, multiple threads are writing data to the same place causing the program to crash. This need to be resolved to at least have a working prototype.
- I believe for multithreading we need to mark explicitly where file/data writing is happening to be able to adjust to make thread-safe
- Double checking that new functions are passing variables in the correct way. The CSE students had this has a slight fear.
- Some helper functions are still not completed (3-6, 8A, 8C, 8D)
- Current completed parts of code are all in separate branches and need to be merged after double checking that variable passing is correct |
|
|
148
|
Fri Mar 25 17:14:01 2022 |
Alex M | Antenna Minimum Length Investigation | The reviewer of the paper we are trying to publish (as well as other external colleagues we showed the paper to) asked about our minimum length we implemented in the loop. Currently, we cut off the length at a minimum of 37.5 cm for each cone (minimum of 75 cm for the full cone). There is an ELOG post from Amy that presented the reasoning for this ( http://radiorm.physics.ohio-state.edu/elog/GENETIS/81 ). We initially thought that we needed a minimum because the antennas were evolving to be small (around and lower than the minimum). We thought that XFdtd was being inaccurate at lengths lower than 37.5 cm, so we set that as a minimum. To see if we can replicate any strange behavior from XFdtd, I simulated antennas at and below our minimum length and generated plots of their antennas responses. The genes are listed below, corresponding to the patterns in the plot. The results do not look unusual (that is, not dissimilar to the other bicones we have generated, including the ARA bicone) and seem to show an improved sensitivity in the upward direction for shorter bicones. The equation we used to arrive at this minimum (f = c/(4L)) might be indicative of a maximum length rather than a minimum.
| Inner radius |
Length |
Quadratic |
Linear |
2.4892,37.5,-0.00142604,0.032832
4.64522,37.5,0.00153863,-0.14004
2.4892,34.5,-0.00142604,0.032832
4.64522,34.5,0.00153863,-0.14004
2.4892,31.5,-0.00142604,0.032832
4.64522,31.5,0.00153863,-0.14004
2.4892,27.5,-0.00142604,0.032832
4.64522,27.5,0.00153863,-0.14004
2.4892,23.5,-0.00142604,0.032832
4.64522,23.5,0.00153863,-0.14004
2.4892,19.5,-0.00142604,0.032832
4.64522,19.5,0.00153863,-0.14004
2.4892,15.5,-0.00142604,0.032832
4.64522,15.5,0.00153863,-0.14004
|
|
|
27
|
Thu Dec 5 14:32:54 2019 |
Julie Rolla | Adding Constraints To-Do List | In my last post, I mentioned (1) our to-do list for adding the ARA hole constraints, and (2) that we've made a mistake upon implementing it. I've attached detailed steps of that to-do list that I've written, as well as an explanation for the correction that is currently being worked on. Please see the pdf attached for an explanation of corrections.
|
|
|
211
|
Tue Apr 11 00:36:49 2023 |
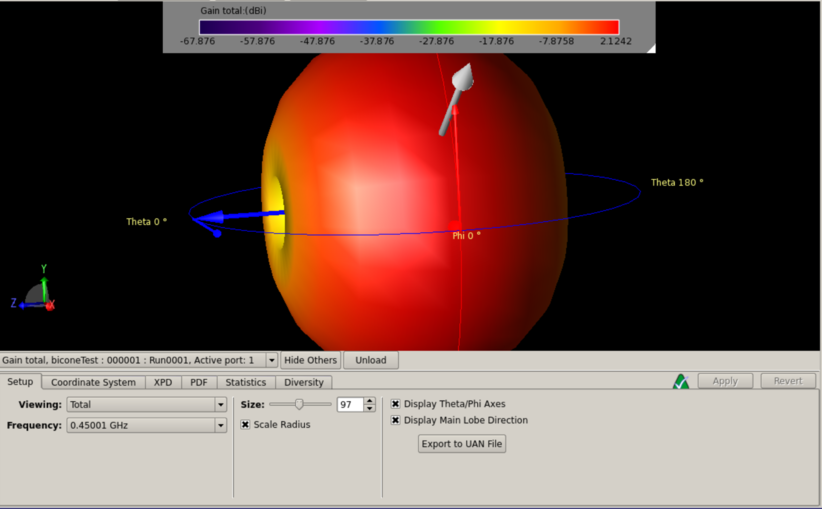
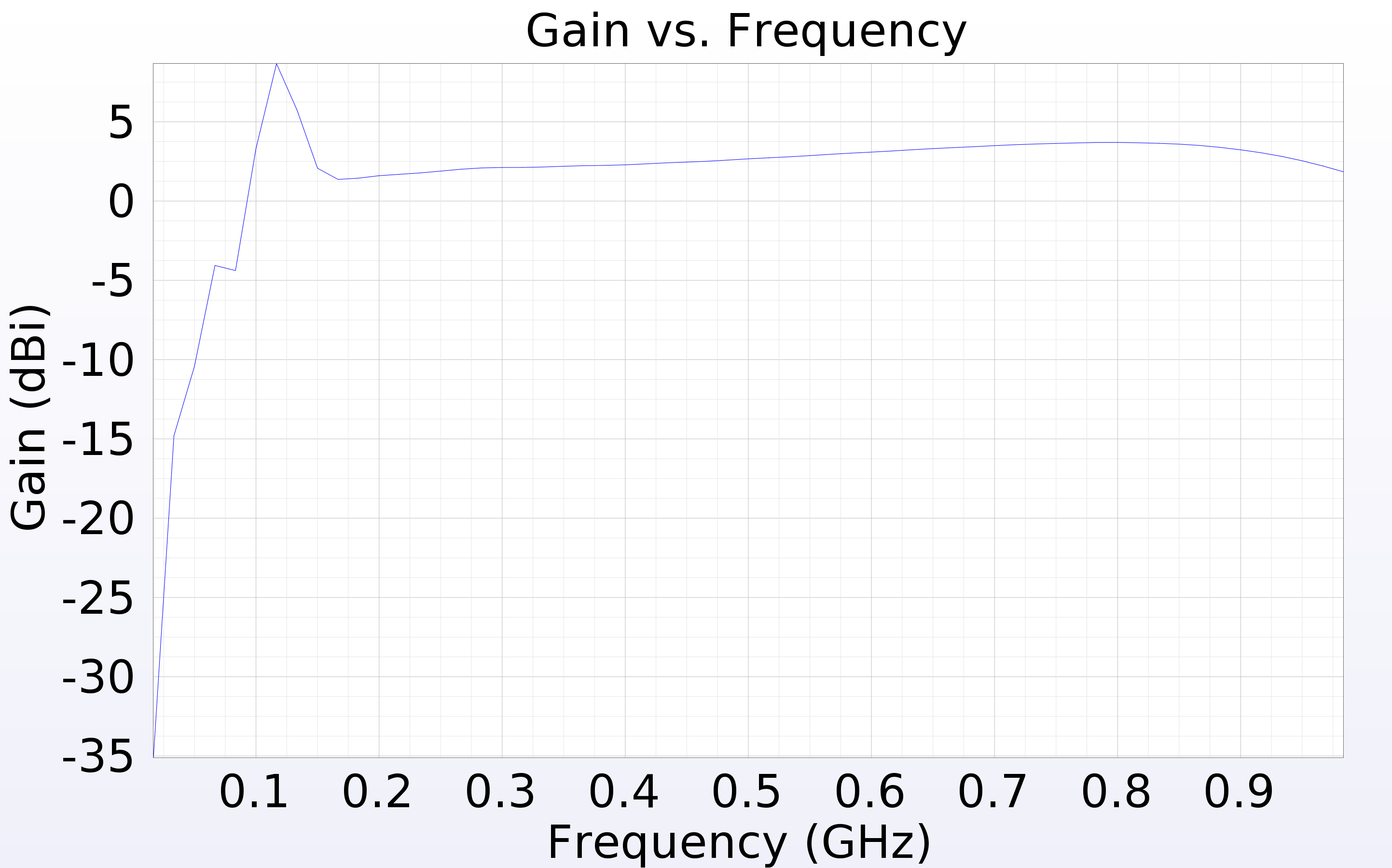
Bryan Reynolds | AREA physics of results plot first look | After some work, I've gotten the physics of results plotting script provided by Dennis working to compare the ARASim outputs from an AREA run to a set of reference data for the ARA bicone (provided by Alex). Among the outputs is a histogram of of events triggered vs cos(theta_nu) (attached here). This is the plot that was previously presented in GENETIS talks in the physics of results discussion.
Here I compared the best individual from the AREA results (Blue- "source_2") that I have on hand (only the ARASim output files from the most recent generation ran are currently saved) with the ARA bicone reference data (Red- "source_1"). Both datasets included a total of 150,000 NNU. Note that the AREA run used was a single frequency test, meaning that the radiation pattern was held constant for all frequencies in the band.
Also attached for reference are the gain pattern of the associated individual from the AREA output and the violin plot of the run producing the data. |
|
|
218
|
Thu May 11 15:57:08 2023 |
Ryan Debolt, Byran Reynolds | AREA Updates | Here is some backlogged information as well as recent updates to our progress on AREA and its optimizations:
4/13/2023
We concluded our initial test of the AREA optimization loop. While analyzing our results, we noticed that most of our runs never reached our benchmarks for performance (chi-squared scores of .25 and .1) and that those that did contain abnormally high amounts of mutation. From previous runs using the GA for the GENETIS loop, we find that mutation and immigration (Note: AREA does not use immigration) usually play a smaller role in overall growth and are simply there to promote diversity. Crossover on the other hand is what handles the bulk of the growth done by the GA. Given our results ran counter to that, we decided to look further into the matter.
Upon looking at the fitness score files, we could see that mutation-created individuals did not perform well in the best run types, but the crossover ones did contain the bulk of good scores. This lines up with expectations. However, closer inspection revealed that these crossover-derived individuals had extremely similar fitness scores, which could indicate elitism in selection methods leading to a lack of diversity in solutions. We then looked at a run that used only crossover with one selection type (roulette), and found every individual to have identical (and poor) scores that were the same up to around the 4th decimal place. This is a strong case that the selection methods used are too elitist to grow a population properly.
We dove into the AREA GA to take a look at the functions doing the selection methods, and discovered several issues potentially causing excess elitism in the GA. It seems that the roulette selection does not behave the way we have used in our other GAs. There appears to be no weighting of individuals; rather, a threshold fitness score is collected and only individuals with a fitness score above the threshold are selected. This causes elitism, as only the most fit individuals are likely to pass this threshold. Tournament was found to be similarly troubled, with a bracket size of ⅓ of the population. This is again very elitist, as it is very large, making it very likely that only the best individuals will be selected. Before we try to further optimize the AREA GA, these selection methods should be addressed and fixed.
For roulette, we propose implementing a similar weighting scheme for individuals as is used in the PAEA GAs that gives preference to the most fit individuals, but still allows others to be selected to encourage diversity of solutions.
For crossover, we propose decreasing the bracket size. This would allow a more diverse range of individuals to be selected through “winning” smaller brackets that would have a lower probability of including top performing individuals.
4/20/23
We (Bryan and Ryan) met to work on improving the selection method functions in the AREA algorithm.
For roulette selection, we added weighting by fitness score, as is done in the other GENETIS GAs, in an effort to prevent it from disproportionately selecting the most fit individuals only to become parents. As a first test, we ran an abbreviated version of the test-loop code, printing the fitness scores of the parents selected by the roulette method. The fix appears to have passed this initial test, as the fitness scores showed more variety and in the case of roulette crossover two unique parents seemed to be selected.
For tournament selection, the only change necessary appeared to be decreasing the bracket size of the tournament(s), which in turn encourages other individuals besides the most fit to be selected as parents, therefore increasing diversity of solutions. We ran out of time to test this change fully, so we will pick up here in our next meeting.
Looking further ahead, we will plan to work on refactoring the AREA algorithm to use the same functions as the other GENETIS GAs wherever possible. In the meantime, this working version can still be used for feature development so that progress is not halted.
5/11/2023
Upon looking at the results of the recent optimizations, our best runs are still taking over 40 generations to reach an optimized score. Given the speed of AraSim, this is still too slow to be considered optimal. Comparing this test loop run to the ones run on by the broader GENETIS GA, our best runs for the AREA GA are worse than the worst runs from those optimizations. As such, we have decided to pause optimizing the AREA GA in favor of adding it to the broader GENETIS one. As of today, we have finished a rough version of the generating function necessary to create new individuals. We have also started progress on the constraining functions and scaling functions that will be necessary to generate the spherical harmonics used to find the gain and phase. Once these functions are complete, we should be able to transplant this GA back into our test loop for AREA and resume optimizations.
|
|
|
151
|
Fri Apr 8 16:06:31 2022 |
Aidan Snyder | AREA - frequency linear dependence correlation test run - 04/05/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- to see if the linear dependence on frequency was properly implemented
- What is different about this run from the last?
- Under closer examination, the previous run of AREA appeared to not linearly depend on frequency, so we recompiled the GA in order to fix this
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Number of neutrinos
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
- Result
- There is a problem with how the GA has assigned the Veff values, resulting in many individuals being assigned zero Veff, however the linear dependence seems to have been implemented successfully
Note: We also ran a run called 20220405fahimi5run1, which was a short test run with a very low amount of individuals and neutrinos which seemed to work fine; as in the linear dependence seemed to work. |
|
|
146
|
Fri Feb 11 16:34:54 2022 |
Aidan Snyder | AREA - frequency linear dependence correlation test run - 01/18/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- to see if the linear dependence on frequency was properly implemented
- What is different about this run from the last?
- attempted to evolve gain pattern as a linear function of frequency
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
|
|
|
153
|
Tue Apr 19 17:46:07 2022 |
Aidan Snyder | AREA - Short run testing stringReplacement2.py - 04/19/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- Check to see if stringReplacement2.py changes were successfull
- What is different about this run from the last?
- added below code in order to create a fitness file inside of each generation to display the fitness scores for just that generation, as opposed to just having one big file full of all the generations.
f2 = open(source + "/" + "gen_{}".format(gen) + "/fitnessFile_gen_{}".format(gen) + ".txt", 'a')
f2.write(','.join(mean_Veff_array) + "\n")
f2.close()
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
|
|
|
190
|
Tue Jan 31 15:31:29 2023 |
Alex M | ARA Bicone Responses | Here we will record where we obtained the ARA bicone antenna response files we use as our baselines. We want to record where we found them/who gave them to us and how they were generated (either through simulation or from actual tests). |
|
|
15
|
Tue Jul 2 13:16:30 2019 |
Julie Rolla | APS slides | The following are links to the 2018 and 2018 APS slides.
2018 APS talk:
https://www.dropbox.com/home/GP_Antennas/APS%20Talk?preview=April+APS+Talk_final.pdf
2019 APS talk:
https://absuploads.aps.org/presentation.cfm?pid=15183 |
|
|
202
|
Mon Mar 20 12:37:38 2023 |
Amy | ANITA/PUEO pictures | Here are some ANITA/PUEO pictures that I received from Christian Mike that might be helpful. I include his descriptions as well.
The one with the black crates in the background is the ANITA 1 and ANITA 2 form factor and the one in the anechoic chamber is the ANITA 3 and ANITA 4 form factor. I dimensioned out an ANITA 3/4 antenna, in CAD space, to give you a better idea of the ridge profile. The ANITA 1/2 CAD models we have, I found, are inaccurate.
Attached are images of the PUEO antenna array geometry. One image annotates the approximate locations of the Vpol feed with the top antenna feed as the origin.
|
|
|
81
|
Thu Jul 2 23:13:05 2020 |
Amy | A useful reference on bicone antennas | I found this datasheet on bicone antennas, that is a useful reference for seeing the relationship between bicone dimensions and bandwidth. From these it looks like the lower end of the band is at about f_low=c/(4*L_E), where L_E is the entire length of the antenna (twice the length of one cone), although we'd expect the peak gain to be where a quarter wavelength is just one of the cones (and the figure may roughly support that).
The attached file comes from this website:
https://www.ramayes.com/Biconical_Antennas.htm
|
|
|
10
|
Tue Mar 5 16:36:51 2019 |
Julie Rolla | 3/5 Task Updates | Tue Mar 5 17:29:02 2019
The things we worked on this week is in bold. Below the bold items is a description of exactly what we took care of.
- Paper:
- Julie added Amy's edits.
- Julie let Kai know he is welcome to edit, too.
- Julie will also keep edits updated as they come in!
- Get loop working-- specifically XF:
- Cade has now officially gotten reasonable outputs for his XF scripts. See the attached images for the XF outputs and associated CAD drawing. It seems to look like a reasonable gain pattern.

- The loop now works until AraSim. We are now getting the following AraSim errors:
- XF_Loop.sh: line 244: ./AraSim: No such file or directory
- XF_Loop.sh: line 244: ./AraSim: No such file or directory
- Julie is going to help Cade make sure that this directory is correct and that it has been compiled! More info to come.
- Test GA (Suren):
- I've been working on creating a gaussian fitness score with which I can test the performance of our genetic algorithm. The idea is as follows: We run the GA and get a generation of individuals. We provide their fitness score by simply taking the values {radius, length, angle} as the {x, y, z} coordinate in a 3D space. We can then use a 3D gaussian to output a value at that location, and use that number as our fitness score. The beauty of this method is that we can then plot the progress of our genetic algorithm each generation as a dot in this 3D space, and we can see it slowly snake to the center. Additionally, we can use more complex fitness scores, like one with a local maxima and a global maxima to see if it get's stuck in the local maxima. Below, I have an image I generated on python of a 2D section contour plot of a 3D gaussian in length-angle space. The dark purple is a lower value and the yellow is a higher value of the gaussian. The lines and dots hand-drawn on top are an idea of what it would look for like to start the GA at the red dot and see it's progress each generation until it reaches the center, shown by the white dots. Running this for our current roulette algorithm would make a very nice GA proof of concept plot for the paper, I believe!
.png.png)
- Gain pattern plots -- Make them 3D (Evelyn) & put on GitHub
- We need to figure out how to make a group Github. We have a lot of Githubs made by people and we need them all accessible from a group account. Right now the old dipole Github and the CalPoly stuff are linked to a group account. We need to find out how to get the Bicone account held by Max, and Evelyn's plotting code all on the group account: https://github.com/gp-antennas.
- Update: Evelyn and Max have contacted Brian to get their accounts linked. It looks like Amy, Brian, and Stephanie (Cal Poly) are the owners.
- Insert LR plot & Gain plot to loop (?)
- Edit bash script
- Update Github
- AraSim plots (Julie and Max) -- see #2 and #3 here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/7 for details.
- Max and Julie have gotten sample data on Owens.
- located at /users/PCON0003/cond0068/ARA/AraSim/branches/AraSimLite/outputs
- Max and Julie have gotten sample programs from Jorge.
- https://github.com/toej93/AraSim_noise_calib/blob/master/readTree.cc
- https://github.com/toej93/AraSim_noise_calib/blob/master/M.readTree
- Julie installed root, and had Max source Julie's thisroot.sh files to run root on Owens.
- Max installed Arasim to his user.
- Next step: run plotting software given by Jorge.
- Update manual with things from Suren's last post here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/8
|
|
|
Draft
|
Mon Nov 8 17:04:30 2021 |
Ethan Fahimi | 11/04/2021 AREA run 2 violin plot | This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation. |
|
|
138
|
Fri Sep 17 13:41:36 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot | This is a plot made from the AREA project with full Arasim implementation. It can be seen that the Veff of any individuals is not what I would consider "good", nor is it really rising, it is quite flat. This is because in this version of AREA, the gain pattern at each frequency is generated differently than each other frequency, there is no correlation. This is known and actively being corrected. This plot is of old data and was just made for two reasons: to make sure that the violin plotting script works for AREA, to display this early form of AREA that has been adapted for full Arasim.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
This plot is further detailed in Julie Rolla's doctorate thesis. |
|
|
140
|
Mon Nov 8 17:27:01 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot | This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
individual 32 in gen 20 and individual 35 in gen 27 look promising (they have Veff > 8) |
|
|
216
|
Tue May 9 18:09:35 2023 |
Alex M | 05/08/23 Pueo Run | Here are the run details for the latest PUEO run we've started. There are still some bugs that need to be worked out, which are detailed on the github. Here is the directory where the data is stored: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_05_08
This run uses 50 individuals per generation with 400k neutrinos per individual at an energy exponent of 19. Only the inner side length and outer sidelength are being evolved (which evolves the height since the opening angle is fixed).
Issues With Part E (Dylan):
The Plots were not automatically created for the first generation. After a quick look through, I found that the fitness score and v effective csv files were being moved before they were copied, so the files didn’t end up in the correct place. Additionally, the errorbars.csv file was not being created, so I made a temporary fix to create a file of 0 error bars to allow for plotting the values while I work on getting the real error values.
Update: I have modified Will’s script for reading the root file outputs, so it can now read them to output a CSV file with the effective volume along with its positive and negative errors. This script does the same and more as the fitnessfunctionPUEO.py, so I will replace it in the loop when there is downtime. Additionally, this script means we don’t need to output a veff.csv file from pueoSim. I will now work on creating a less “janky” version of my pueoSim change for reading in the gain files, so I can get Will to officially add it to pueoSim and we can pull any updates in the future.
Update, Implementing the new script into Part E:
The rootAnalysis.py script is very picky with environment variables. So, first I had to load and then unload a version of python to make sure the default version is loaded. Then, I moved the sourcing of set_plotting_env.sh to later in the script to avoid having an alternate version of root sourced while running (this would cause errors). Then I source the environment for pueosim (path hardcoded, change when loop isn’t running) and loop through the rootAnalysis.py program for each individual in the generation. This will output a ${gen}_fitnessScores.csv and a ${gen}_errorBars.csv in the run directory for plotting script to use. Because it outputs these automatically, I commented out the old methods we used to create these files and removed the temporary error bar fix as well.
Another Update:
We paused the loop. I changed the hardcoded parts, and we pushed the new updates to the github. I will make comments on the changes later today.
Wednesday:
It seems like the loop is still not creating the csv files or plots for generation 1 or 2. Alex gave me the error message. The python script rootAnalysis.py had an issue with the include Geoid.h line. This was caused by sourcing the wrong things while trying to get the script working. I found that I only changed my local pueoSim environment to include the fftw installation to the $PATH and $LD_LIBRARY_PATH variables. So, I changed /fs/ess/PAS1960/buildingPueoSim/set_env.sh as well. Also, I found that the include statements in rootAnalysis.py were missing the PAS1960 portion of the path. After fixing these, and adding a missing ‘/’ to the output file path, I manually ran the rootAnalysis.py program for the 2023_05_08 run, mimicking the portion of the bash script that runs it in Part_E. It outputted the correct files in the correct format. I then ran the FScorePlot script. The median line for the ViolinPlot seemed wrong, and the FScorePlot was too scrunched with a y start of 0; I will need to change these.
Plotting Update (Dylan):
Ok, plots still aren’t being made, and I’m not running the loop, so I don’t have access to the error messages, so I’ll go through everything one by one. For Veff_Plots, this isn’t being made as there are currently no vEffectives.csv. This isn’t a big deal, as it’s just a copy of FScorePlot, but I added the creation of vEffectives.csv to rootAnalysis.py anyway. Next, I noticed that runData.csv, which is required for a couple of the plots was not being made. I looked, and the version of gensData.py had the old values for the skiprows when reading in the fitnessScores and generationDNA files, something that was overlooked when we merged the version I was developing with the version Alex was developing on. I can’t access the directory or the file, I think this is why it was overlooked in the merge. I created a temporary PUEO version in Antenna_Performance_Metric (I’m also fine with it just living here permanently).
Also, the csv files are now in the GenerationData/ directory in the run directory? This affects every plotting script and they will not work without changes. I don’t know why this was changed or if this change is permanent?
Next, ok apparently, VariablePlots isn’t even a file in the loop? We really need to use GitHub better.
I moved my local version of the program into the loop and it ran fine manually.
I am forsaking the Generation_Data directory, so I changed the rainbowPlotter and dataConverter scripts to read in a location to match all the other plotting scripts. They both also ran fine manually.
Note: The rainbowPlotter script should be able to create a gradient from the minimum fitness score to the maximum fitness score. (currently, the range is hardcoded and this can cause basically a mono-color graph.)
FScorePlot should be good
The bash script was inputting 5 variables into colorplots when the pueo version only takes in 4 (this is also something that was already fixed before the merge.) It also requires the vEffective.csv files. After copying over the fitnessScore.csv files to vEffective.csv files, it ran fine. (The vEffective.csv files will be made automatically by rootAnalysis.py in the future) |
|