| |
ID |
Date |
Author |
Subject |
|
|
8
|
Fri Feb 22 17:09:28 2019 |
Suren Gourapura | Updates that need to be added to the manual | We are redesigning the way we simulate antennas in our loop. To do this, we changed our simulationPEC macro skeletons and our output skeleton.
To make this easier, we changed the way we name the files, from i.uan where i is the simulation number, to i_j.uan where i is the antenna and j is the frequency.
We also rewrote XFintoARA.py so it's clean. Note that before there were comments from a previous author claiming it was VERY questionably written. This has now been cleaned up. We also changed the bash script relevant to the above changes.
This change needs to be added to the manual, and until we update this update, we have not done it.
|
|
|
28
|
Thu Dec 5 16:58:55 2019 |
Julie Rolla | Updated To-Do List: | Updated To-Do List:
- Add ARA constants in plotting code: ie add Veff, theta, R for ARA. This will sit as a constant line for us to compare each evolving parameter to. For example, in the plot Veff vs Generation, we will see a constant line for ARA's Veff to see how it is evolving in time.
TASK 1: MAKE Veff PLOT. Note: One of our variables in this run is Energy. Unfortunately, we care about how Veff changes with Energy for ARAactualBicone. This means that before plotting "Veff versus Generation" we need to generate Veff for ARAactualBicone at the energy we are assigning the run.
STEP 1: Make plotting code to plot "Veff vs Generation".
- You can follow the way we make LRPlot.py.
- However, in this case, we will need to take the files /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Ouputs/$RunName/AraOut${gen}_${i}.txt -- ie the AraSim outputs for each individual in ALL generations. ie Gen 0 - ${gen} -- INCLUDING the one we created in Gen 1 for ARAactualBicone -- and plot Veff from those.
- We also want to plot error bars with Veff. The error bars for each are labeled "and Veff (water eq)" at the bottom of these AraSim output files we are already reading in for Veff.
- Note, to know how to read in Veff, you can look at how we are reading that *exact* line from the AraSim output files in on the fitnessFunction_ARA.cpp code in the Antenna_Performance_Metric directory, since we are doing it there.
- Note that since we are plotting "Veff vs Generation", we will either have to read in all the files AraOut${gen}_${i}.txt (up to how may ${gen} we are in) every time -- including the ARAactualBicone one-- or we can append them all to one .txt file. It's your choice.
- Make sure you label the units for each axis every time, and that you have a key! If not, Amy will hate the plot!
STEP2: Add Veff for ARAactualBicone as a constat in the newly made "Veff vs Gen" plotting code. Remember, as I mentioned above, we must first put the ARAactualBicone through AraSim to see Veff at the same energy as the rest of the individuals. Here's what to do:
- Edit the Bash script to submit a job that runs the gain patterns of ARAactualBicone through AraSim in the initial gen (gen 0)!
- There's a current file that exits for ARAactualBicone that has the gain patterns of the current antenna, in the correct format for AraSim to run. In the bash script (XF_Loop.sh) we need to create ONE more AraSim job submission for this antenna (as a comparison to what we see with our genetically evolved ones) just in gen 0 (the first generation).
- This current AraSim input file for ARAactualBicone exists in BiconeEvolutionOSC/AraSim/Ara_Bicone6in_output.txt
- This needs to be the input for our .job script.
- Add lines to your newly made "Veff vs Gen" plotting code that reads in the Veff output for this ARAactualBicone AraSim output file (whatever you've chosen to name it when you wrote it into the job script.
STEP 3: Move the ARAactualBicone output file into the $Run_Name directory so it is not overwritten ever.
- We will want to make sure this is not overwritten after the run ends, and we start a new one.
- We will also be using it for plotting "Veff vs Generation".
STEP 4: Correct the "wait" command for the AraSim job submissions.
- The "wait" command, on the first iteration, will have to wait for $NPOP+1 now-- not $NPOP.
TASK 2: MAKE "Theta vs Gen" Plot.
STEP 1: Make plotting code to plot "Theta vs Generation".
- You can follow the way we make LRPlot.py; however, MAKE SURE YOU LABEL AXIS WITH UNITS, AND HAVE A KEY.
- This will be exactly the same as this other code, except you will need to:
- Read in theta from your generation DNA files
- Add the ARAactualBicone constant to the code as a reference point
TASK 3: Fix "L vs Gen" and "R vs Gen" plot.
STEP 1: Correct both plots to have labeled the units for each axis every time, and a key. This is plotted in the "LRPlot.py" code.
STEP 2: Fix our 'R' values. I do believe we are plotting the incorrect "R". Here is what I think is happening:

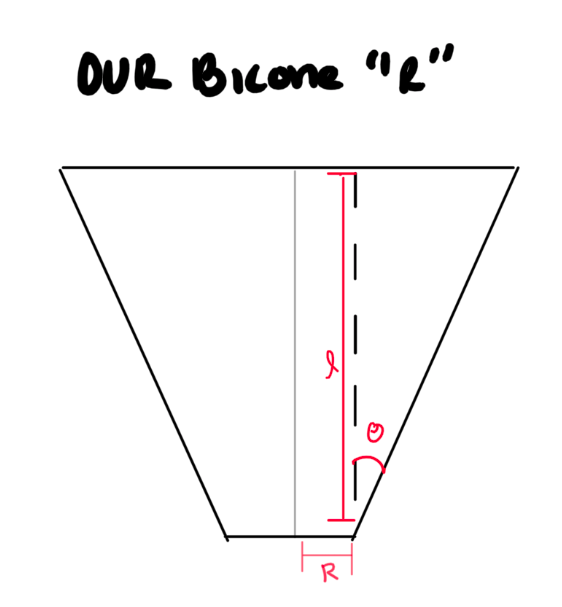
- We need to double-check and see what ARA defines "R" as on their current antennas.
- Check with Amy?
- What does Brian Clark's thesis say?
- Can you find a paper that shows a picture of how they define it?
- If we are wrong, this means we need to change the "R vs Gen" part in the "LRPlot.py" code so that we:
- Read in R, L, and Theta for each generation.
- See how this is already done for reading in R, and repeat it for L and theta.
- Do trig to calculate the same "R" as ARAactualBicone defines. See picture below!
- Store that newly calculated "R" as an array, and plot THAT.
- Don't forget to add the ARAactualBicone Radius as a constant line.
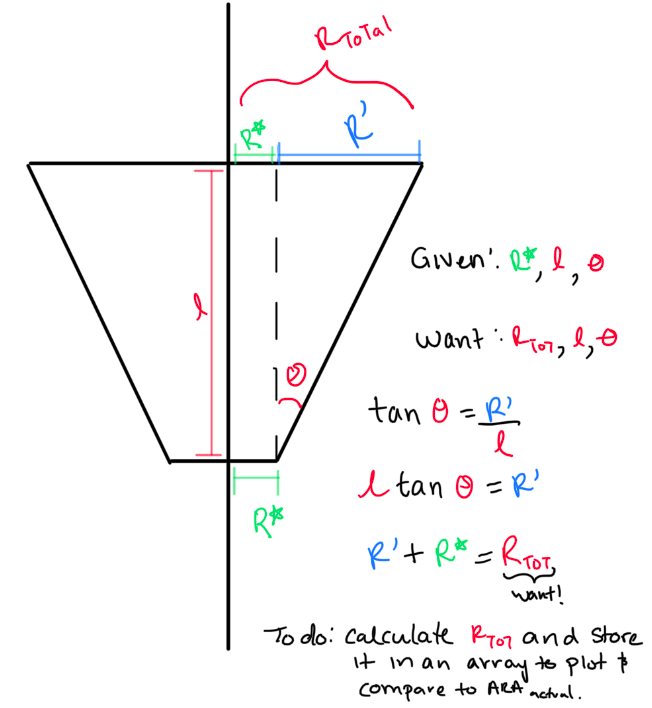
TASK 4: See PDF for other items! |
|
|
7
|
Fri Feb 22 15:46:33 2019 |
Julie Rolla | Updated To-Do List | We are getting to the stage where we would like to verify each step. It has been requested by Amy that we verify them visually. Here are the plots we need to make.
- Add plotting for L & R parameters (note that we already have similar software to this from the dipole check. You can find this in the GitHub from the dipole test here: https://github.com/hchasan/XF-Scripts). This should be between A, and B in the loop drawing on the manual. (Done)
- Plot the distribution of theta and phi of incoming signal. This should be after D in the loop drawing on the manual.
- Plot the distribution in cartesian coordinates of the neutrino interaction position. Maybe we can also plot the distance between the interaction point and the antenna. This should be after D in the loop drawing on the manual.
Finally, as a tasks not to do with plotting, we need to make sure we don't lose any data as we go. We want to make sure that we keep tagged or labelled which individuals survive, and save the data flagged for those as we go. |
|
|
175
|
Fri Aug 5 09:59:07 2022 |
Dennis Calderon | Updated Effective Volumes and Errors (Paper Run and Curved Sides) | Update to previous ELOG post for Effective Volumes and Errors for AraSim simulations for antennas from Paper_Run and Curved_Sides.
AraSim version compiled ~11/2021 and the (old) GENETIS version
Here are the column definitions:
- The first column indicates the antenna being described. For example, XF_model_21_20 was the 20th individual in the 21st generation for the run discussed in the paper. Curved_side_1 was the best performing individual from the curved run done in February 2022. Curved_Side_1_Quad_Zero indicates the curved individual with its quadratic term set to 0. Curved_Side_1_Straightened indicates the curved individual with the quadratic term set to zero and the linear term modified so as to keep the outer radius the same as it originally was (that is, for Curved_Side_1).
- The next three columns are the measurements for that individual.
- The first one idicates the mean effective volume of the runs done for that individual. There were ~300 runs, each of 10,000 neutrinos.
- The second column indicates the standard deviation of the mean (the previous column). This was measured by taking the standard deviation of the effective volumes (the line that says test Veff(ice) in the AraOut file) from all 300 runs (that is, Sigma(mean - x_i)/Sqrt(300)).
- example of line:
- Veff(ice) : 5.25337e+09 m3sr, 5.25337 km3sr
- The final column indicates the average of the errors on the effective volume measurements from the AraOut files (the line that Veff(water eq.)) divided by Sqrt(300) for each of the 300 jobs (that is, Sigma(x_i/Sqrt(300))/300).
- example of line:
- And Veff(water eq.) error plus : 0.552722 and error minus : 0.552722
Note: Previously, the best curved individual (according to the run data) had been excluded (instead, we had the 2nd-6th best individuals). This has been corrected, with the order corrected.
Also find attached a file called full_resimulate_DNA.csv . It contains the DNA for the top five curved individuals, the DNA for those same individuals with their quadratic term set to 0, and the DNA for those individuals with the quadratic term set to zero and the linear term modified to maintain the same outer radius as in the original run.
Finally, the three images show the original best curved bicone (Curved_Side_1), the best curved bicone when its quadratic gene was set to 0 (Curved_Side_1_Quad_Zero), and the best curved bicone when its quadratic gene was set to 0 *and* its linear gene was changed to keep the same outer radius (Curved_Side_1_Straightened).
We have chosen XF_Model_23_08 to be the one actually built for testing. This was decided based on the data presented here. XF_Model_23_08 was the highest scoring individual in the paper run. Because the paper data was generated using the old version of AraSim, it was decided that using those results would make the most sense for which antenna to build. This would also mean that we can compare directly to the best individual in the paper. This individual was chosen over the curved sided inviduals because it has a higher fitness score, both when using the old version of AraSim and the new version. |
|
|
Draft
|
Mon Oct 5 17:34:32 2020 |
Leo Deer | Update Monday 10/5/2020 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
Finish up mutation function and then make sure it functions properly, currently our added functions all compile but we haven't tested if they are functioning as intended yet. Also have some questions about handling the standard deviations in mustations but first we want to get it functioning before applying more tweaks |
| Eliot |
Worked on learning about 3D plotting in python. Made a simple surface plot that can be expanded upon for our purposes. |
|
| Leo |
Fixed the averge Fscore Plot so that it connects the mean values with a line instead of making them discrete points. Then I made Kai's plot suggestion. For that I copied the current FScore plotting software and made some edits so that it takes in 2 runs and plots the fitness scores of both in either red or green along with the average fitness score of both. I'll attach images of what these 2 plots look like. |
For Friday I'm going to look over the other plotting suggestions but most likely begin on fixing the random seperation in the RLTS plots. |
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
109
|
Fri Oct 2 17:11:25 2020 |
Ethan Fahimi | Update Friday 10/2/2020 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
Today I started working on improving the plots. I started with plotting the average fitness score for each generation on top of all the individuals data points. I made the average fitness score a star with higher contrast so it will stand out. Here is the plot of the first 26 generations of the "Machtay_20200914_Asymmetric_50_Individuals" run with the new averages. |
On monday I'm going to try and create the plot of 2 runs overlaid on top of eachother as Kai suggested. |
| Evelyn |
|
|
| Ryan |
Worked with Ben and Alex P. on making the Bicone function modular on our copy. We finished writing a new roulette selection function, tournament selection function, and reproduction function. |
Finish writing the mutation and crossover functions. After these are complete apply these functions in place of the existing function that is wrong and test to make sure the results are not producing errors. |
| Ben |
|
|
| Ethan |
Updated the AREA code with Alex M's help to work using SLURM. Began a trial run to see if our updates work. |
Begin making changes that allow the job to run faster as well as investigate what is causing some individuals to have fitness scores beyond what should be physically possible. |
|
|
|
107
|
Wed Sep 9 17:28:54 2020 |
Alex M | Update 9/9/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I've been working on readying the loop for the transition to slurm. OSC is switching from its current job managing software (torque/moab) to slurm, which uses some different commands but is functionally similar. Since we have places where jobs are called in the script, I've been changing them (with a backup copy of those files that we know works on torque).
Right now I'm testing all of the changes I've made on the space OSC has set aside for playing with slurm. I have changes in all of the places I think they belong, but I need to make sure there aren't typos/different commands from what I've used.
|
I'll keep testing the changes I've made for slurm. I'll also try to organize some of the dfferences I've found in using slurm vs torque (for example, requesting an interactive job) so that everyone can see them and be able to find out what to use when OSC makes the switch (Pitzer changes on the 22nd, Owens is in November I think). |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
127
|
Tue Jan 26 17:17:41 2021 |
Ethan Fahimi | Tuesday Updates 01/26/2021 |
| Ethan F |
Continued implementing some solutions the OSC helpdesk gave me for fixing the unwanted extensions on the job output files. |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
219
|
Mon May 22 20:34:59 2023 |
Amy | Things to do before starting new PUEO run from 5/22/23 meeting | Here is a list of things we agreed to do before starting a new PUEO run. Feel free to add notes here as things are done and we will revisit the list next week.
Change range of the height variable to 0.75-1.75m
Start with all bad individuals so that we can be sure and see growth - all at 0.75m?
Make sure we are using the ratios in the GA that we think we are
If we are using reproduction, are we combining results when individuals are resimulated to reduce errors? I made a program to accomplish this and created a pull request on the PUEO Github (Dylan)
Investigate whether there are obvious places to speed up PUEOSim
Consolidate the GA to one branch
Understand what is happening with cross-pol, make sure we're not gettting Veff=0 when using XF-generated cross-pol response
Edit: Have the xmacros use the realized gain instead of the idealized gain, which allows for the design to build in matching
|
|
|
233
|
Mon Jul 24 15:51:28 2023 |
Ryan Debolt | Test Loop runs that need done. | |
Types of runs I need (May need some other people to help me run these):
-
Optimization runs
-
Non-error
-
Search over combinations of selection methods and genetic operators
-
Fix Sigma at 10%.
-
Do this for 10 runs for each
-
Re-run the best run for 100 tests to see if the results agree with 10 test results
-
This will be the main talking point
-
Save the plot for the best combination (proxy and metric)
-
Error
-
Re-run the best run for 100 tests with 3 different error amounts (0.1, 0.2, 0.3)
-
We will compare these results to the non-error run to talk about how errors may affect consistency/speed.
-
Save an example plot of an error test for each error (both metric and proxy score)
-
Population
-
Re-run the best run for 100 tests with 3 different population sizes (100, 500, 1000)
-
We will compare these results to the non-error run to talk about how population size affects consistency/speed.
-
Save an example plot of a population test for each size (just proxy score)
-
Demonstration runs (single runs, fitness score, no error population 100)
-
Crossover only
-
Mutation only
-
Reproduction only
-
Injection only
|
|
|
241
|
Mon Oct 7 15:35:01 2024 |
Dylan Wells | Template Run Results Slide Deck | Copy this slide deck to use when presenting on run updates!
https://docs.google.com/presentation/d/1Tk-B1QbFTP_5pQZovfn0_wbTswZTmOrJURpi374xJWo/edit?usp=sharing |
|
|
21
|
Wed Nov 20 15:05:14 2019 |
Julie Rolla | Task update 11/20/19 | Today I spent some time looking at task 1 from the update here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/20. Below are my updates.
Task-- Fix error at fitness function:
Currently, the loop stops at the fitness function calculation, and aborts (program fitnessFunction_ARA.cpp). The following are the things I have tried.
1.Run this outside of the loop: Julie and Alex did this 11/19/19. It gave a segmentation fault. UPDATE: HERE'S WHAT I DID TO CHECK THIS TODAY:
(a) Checked using cout: I used some cout commands to print sections looking for errors. It seems we were not properly giving it arguments when running it outside of the loop. Since it's running outside of the loop, these variables are not initialized. Thus, to run you need to type: ./fitnessFunction.exe $NPOP $AraOut_i.txt
- For example: ./fitnessFunction.exe 1 AraOut_1.txt
Status: The output file is verified! Running this way does properly generate our output file named fitnessScore.csv. I've double-checked this file, and it is properly being filled. At the moment it is giving a Veff of 0, thus no valid fitness score; however, this should be fixed automatically once we get real outputs from AraSim without a Veff of 0.
2. Check file location: See if fitnessScore.csv is being located in the correct place when the genetic algorithm initializes the next generation.
(a) Searched bash script for the final location of fitnessScores.cvs:
In lines 298, 299 of the bash script, it is doing the following
cp fitnessScores.csv "$WorkingDir"/Run_Outputs/$RunName/0_fitnessScores.csv
mv fitnessScores.csv "$WorkingDir"
This is taking the .csv output generated from fitnessFunction_ARA.cpp, and is moving it to the run name directory and renaming it based on the generation -- ie first gen is 0_fitnessScores.csv-- so that data isn't overwritten. Then, for some reason, it moves the original copy -- ie fitnessScores.csv-- to the location /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop
In line 374 we then run our genetic algorithm again.
./roulette_algorithm.exe cont $NPOP
This does not seem to take in the input file as an argument.
(b) Checked roulette_algorithm.cpp for the location it accesses fitnessScores.csv:
I checked if it was hard-coded into the roulette_algorithm.cpp script. I found the following lines.
// Now we need to read the fitness scores:
ifstream fitnessScores;
fitnessScores.open("fitnessScores.csv");
string fitnessRead[NPOP+2];
for(int i=0;i<(NPOP+2);i++)
{
getline(fitnessScores,fitnessRead[i]);
if(i>=2)
{
fitness[i-2] = atof(fitnessRead[i].c_str());
if (fitness[i-2]<0)
{
fitness[i-2] = 0; // If the fitness score is less than 0, we set it to 0 to not throw things off.
}
}
}
fitnessScores.close();
}
This means that our script roulette_algorithm.cpp is grabbing the fitnessScores.csv from its current directory-- which is accurate due to the line (mv fitnessScores.csv "$WorkingDir") referenced above in the bash script. Something we could verify here is to make sure this function is properly working by having it cout elements from the fitnessScores.csv within this function and to run the roulette algorithm separately. We could do this now, however, we don't have real outputs in fitnessScores.csv anyway, since we are getting a veff=0 for AraSim temporarily.
In the meantime, I've added the following line after section E (the section that runs fitnessFunction_ARA.cpp, and generates/moves/copies fitnessScore.csv):
echo 'Congrats on getting a fitness score!'
This should check to be sure that it runs until after this point. I've also added the following line after section E (section that plots the fitness score versus gen):
echo 'Congrats on getting some nice plots!'
This should check to be sure that it runs until after this point. Note that I have added these lines to both our original bash script, and the one Alex M. is editing to turn every section into a function. I have added these lines to the .sh scripts containing the functions to be sure the function scripts for these tasks ran completely. Once we have real AraSim outputs, we should be able to create some cout lines in the instructions highlighted in yellow above.
3. Look at how it's calculating the fitness score.
I have not checked this yet. However, I do not think it is likely this is the issue. I will check later though. |
|
|
13
|
Mon Jun 24 13:49:49 2019 |
Julie Rolla | Summer update | Right now the following people are working on this project:
Julie Rolla, Cade Sbrocco, Scott Janse, Alex Patton, Eliot Ferstl, Abassi Brown, Evelyn Shank, Sophie Dunlap
Today we met for our first working meeting. Scott, Alex, and Eliot were asked to speed up and optimize AraSim. Sophie and Evelyn are working on editing the .csv file with the beam patterns coming out of XF. Currently it's written in a different format than AraSim takes in. Abassi is helping Evelyn and Sophie by giving them a sample program helping reorganize the cells in a .csv file. Since there's confusion on how to optimize AraSim, we've started dissecting the software so we can understand how it works. The following are questions on the setup.txt document.
Setup.txt
What is "timestep"? Could we increase the timestep? Is this timestep big or small compared to 2 GHz?
Answer: Timestep is the unit of discrete time the computer operates at. The timestep by default is set to 2 GHz, or 5 * 10^-10 seconds. We might be able to make up some cheap time by lowering that but would have to check if it makes the data less accurate.
What is "TRIG_WINDOW"? Is this a "coincidence"? Meaning if this signal is picked up within a certain time between stations it's considered the same event?
What is "V_MIMIC_MODE"? Comments in Setup.CC says "// default : 0 - write out all chs where global triggered bin is center of the window"
Answer: Not sure exactly what it does but we can't change it from default as the other values are for the test bed mode only.
Random Question from Setup.txt, but relevant for other things. What is "testbed mode"? Answer: The "testbed" is the original test station only 40m under. This mode is for if we do detector=3 in the first part of the setup.txt file; thus, we won't use this.
What's the difference between "DETECTOR" and "DETECTOR_SIMULATION"? We found "DETECTOR_STATION" in Settings.h. The comments say "// for DETECTOR=4, indicates the single station to be simulated // 0 = testbed, 1 = A1, 2 = A2, 3 = A3"
In this file, there's an ANTENNA_MODE variable. You can look at what it does in Settings.h. In this file, it says that you can decide whether to use a bicone antenna response or "a different response for the top vpol antennas". How does this function work? What does it mean by "top vpol antennas"? Someone should look up the ARA antennas, and look into this function.
Our goal is to meet and work as a group twice a week. Collaboration in work should help us progress faster.
|
|
|
229
|
Thu Jul 6 13:12:04 2023 |
Jack Tillman | Straightened Sides AraSim Results | After discretizing and straightening the sides of the curved antenna that performed the best (Generation 13, individual 84) and running the antenna through AraSim, the following results and physics plots were generated. |
|
|
14
|
Mon Jul 1 13:45:49 2019 |
Alex Patton | Status on optimizing AraSim | We ran valgrind (a profiler) on AraSim to find out where it was spending most of its time, we ran it with callgrind to give an output file of the time and opened that file with qcashegrind to display it in a legible format. This let us know that the function Param_RE_Tterm_approx was taking up about 75% of the time because it was being called hundreds of millions of times. Now more specifically the standard math functions pow() and exp() were being called in this function every time and took up about 44% and 20% respectively. They were called about the same number of times as Param_RE_Tterm_approx but they are only called in the else block in an if-else statement.
JULY 31st Update:
I used the C++ library future and set up iterations of a loop to run asynchronously as they do not rely on each other's calculations. This launches a seperate thread whenever I call one. The struggle with this is that launching a thread and getting its results takes time as well, so there had to be enough calulations in a thread in order for it to make up any time. The purpose of the function GetVm_FarField_Tarray is to create arrays of doubles called Earray and Tarray and those are the only values that get altered in this function. Tarray is created with simple calculations so I left that out of threads but making Earray calls the other function that takes up so much time.
I edited the files signal.cc and signal.hh to work with 4 threads and ran 10,000 neutrinos with 4 cores. The cpu time was 1:48:41 but the real walltime it used only took 00:54:40. The time these tests for 10,000 neutrinos with base AraSim normally take anywhere from 1:35:00 to 1:45:00 or in that range. It should still work with less threads available it would just run slower and probably at the cpu time, which might be slightly slower than base AraSim but as soon as more threads are available it runs much faster as we see here. I still need to check for the accuracy of these edits, the number passed is in a reasonable range, but I will need to investigate further by giving the exact same neutrinos to both programs and making sure their outputs are identical.
I also have some further ideas to speed it up even more. Currently every iteration of the loop calls a thread but what would be faster would be to either
A: Set it up so it can call more threads at a time and thus go faster, which would be an easy edit but only work if more cores are given
B: Set it up so the current threads run more than one iteration at a time, I've started experimenting with this but having trouble making sure that everything involving pointers stays safe, but the benefit of this is that the threads would get called and joined significantly less and would cause a great speed increase without adding more threads.
All these developments are new after me spending a while trying many different ways to speed it up or run parts in parallel like this, so even if this isn't accurate or valid I believe I would be able to make it so with more time and work.
Attached I have a zip that contains the two files I edited as well as an output file from my first test. I will continue to test and improve this but am unable to work on this again until August 19th so I wanted to share my current status before I am unavailable. |
|
|
129
|
Thu Mar 11 15:41:52 2021 |
Alex M | Running The Loop Updated | Because of issues we were having with XF when running at the command line, we've shifted to running through a Pitzer VDI (see here: https://ondemand.osc.edu/pun/sys/dashboard/batch_connect/sys/bc_desktop/vdi/session_contexts/new).
The VDI will give you a mirror of a desktop running on OSC's side, so things like XF will run much faster. You can open a terminal by clicking the black box at the bottom of the screen and then run the loop like normal (./Asym_XF_Loop.sh). This has the added benefit of making it possible to exit out of your browser, close your laptop, or turn off your computer without interrupting the run, since the process is running on OSC's side rather than piping to your computer. |
|
|
141
|
Fri Feb 4 16:10:32 2022 |
Julie Rolla | Run Log Template | For PAEA Algorithm:
Part I: Complete as soon as the run starts
Run details: Please answer all of the questions below!
- Run Type
- Answer here whether or not it's AREA (Gain pattern evolution) or PAEA (Bicone evolution)
- Run Date
- Run Name
- Parameters evolved
- Why are we doing this run?
- Add answer here about what we are testing
- What is different about this run from the last?
- Add answer here with info on what we are doing differently from the last run
- Example: We are testing a new operator, we are changing the percentages of each selection method used, etc.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Add answer here (say N/A if this is an AREA run)
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- Selection methods used (% of each):
- Are we using the database?
- Add answer here. This can be found in the main bash script within the variables section. (Answer N/A for AREA run)
Please upload the text file with all run details before closing this!
Part II: Complete as soon as the run ends
Results: Once this run completes, please upload the plot(s) to this post as an attachment, as well as a general explanation of the results.
- Summary and comments on results
- Add thoughts on what we should do next here!
- Example: "It seems like we are seeing minimal evolution. We should take a step back and try to see why we don't see improvement. Once we trouble shoot, we can start a new run to investigate."
- Example 2: "Run looks good! Our best individual is in generation #12. Attached are the CAD drawings of that individual."
**Upload all plots, run parameter text files (file that has run settings saved), CAD models of best indivduals, etc**
For AREA Algorithm:
Part I: Complete as soon as the run starts
Run details: Please answer all of the questions below!
- Run Type
- Answer here whether or not it's AREA (Gain pattern evolution) or PAEA (Bicone evolution)
- Run Date
- Run Name
- Parameters evolved
- Why are we doing this run?
- Add answer here about what we are testing
- What is different about this run from the last?
- Add answer here with info on what we are doing differently from the last run
- Example: We are testing a new operator, we are changing the percentages of each selection method used, etc.
- Single frequency run or run with broadband frequency dependence:
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- Selection methods used (% of each):
- Any other things to note?
Please upload the text file with all run details before closing this!
Part II: Complete as soon as the run ends
Results: Once this run completes, please upload the plot(s) to this post as an attachment, as well as a general explanation of the results.
- Summary and comments on results
- Add thoughts on what we should do next here!
- Example: "It seems like we are seeing minimal evolution. We should take a step back and try to see why we don't see improvement. Once we trouble shoot, we can start a new run to investigate."
- Example 2: "Run looks good! Our best individual is in generation #12. Attached are the CAD drawings of that individual."
**Upload all plots, run parameter text files (file that has run settings saved), gain patterns of the two best and two worst individuals, etc**
|
|
|
163
|
Tue Jun 21 12:02:23 2022 |
Alex M | Run Details: Machtay_20201023_300K_Nus_50_Individuals | This run was started on 10/23/2020. The purpose was to attempt to demonstrate evolution by beginning from 50 identical individuals in the initial generation (which had previously produced a low score).
This run was the first time we removed the penalty on fitness scores for antennas which exceeded the borehole radius. It was also the first time we increased the number of neutrinos thrown up to 300,000. 50 fully asymmetric individuals were evolved over 25 generations.
There was a 50/50 split for roulette/tournament selection and 75%/10%/15% for crossover/reproduction/immigration. While the evolution was somewhat flat, we do believe we demonstrated evolution because the average score rose as the run evolved.
(Note that the final generation was interrupted but the plot was still made, hence the drop to 0 for all scores on the plot) |
|
|
168
|
Tue Jun 28 11:40:32 2022 |
Alex M | Run Details: Machtay_20200911_Symmetric | This was a symmetric run that began on 9/11/2020. This run used fewer neutrinos to determine how significant the number would be on the performance of the evolution. 30,000 neutrinos were used per individual.
10 individuals were evolved over 35 generations, using 100% roulette selection and the old mutation algorithm, where genes had a 40% chance of mutating. Mutations changed the gene by reselecting the value from a guassian with a mean and sigma equalt to the mean and sigma of that gene's value in the generation.
|
|
|
167
|
Tue Jun 28 11:35:15 2022 |
Alex M | Run Details: Machtay_20200831_Asym_Length_and_Angle | This run was conducted beginning on August 31, 2020.
Both the angle and length were asymmetric for the two cones. It was run with 10 individuals over 42 generations using 150,000 neutrinos. A penalty was still implemented on bicones exceeding the borehole width.
As this was before the substantial rewrite of the genetic algorithm, 100% of selection was done using roulette. Genes had a 40% chance to "mutate," which was done by selecting a new value from a guassian, with a mean and sigma equal to the mean and standard deviation of that gene's value in that generation.
|
|