| ID |
Date |
Author |
Subject |
Project |
|
50
|
Wed Jun 12 12:10:05 2024 |
Jacob Weiler | How to install AraSim on OSC | Software | # Installing AraSim on OSC
Readding this because I realized it was deleted when I went looking for it 
Quick Links:
- https://github.com/ara-software/AraSim # AraSim github repo (bottom has installation instructions that are sort of right)
- Once AraSim is downloaded: AraSim/UserGuideTex/AraSimGuide.pdf (manual for AraSim) might have to be downloaded if you can't view pdf's where you write code
Step 1:
We need to add in the dependancies. AraSim needs multiple different packages to be able to run correctly. The easiest way on OSC to get these without a headache is to add the following to you .bashrc for your user.
cvmfs () {
module load gnu/4.8.5
export CC=`which gcc`
export CXX=`which g++`
if [ $# -eq 0 ]; then
local version="trunk"
elif [ $# -eq 1 ]; then
local version=$1
else
echo "cvmfs: takes up to 1 argument, the version to use"
return 1
fi
echo "Loading cvmfs for AraSim"
echo "Using /cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
source "/cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
#export JUPYTER_CONFIG_DIR=$HOME/.jupyter
#export JUPYTER_PATH=$HOME/.local/share/jupyter
#export PYTHONPATH=/users/PAS0654/alansalgo1/.local/bin:/users/PAS0654/alansalgo1/.local/bin/pyrex:$PYTHONPATH
}
If you want to view my bashrc
- /users/PAS1977/jacobweiler/.bashrc
Reload .bashrc
- source ~/.bashrc
Step 2:
Go to directory that you want to put AraSim and type:
- git clone https://github.com/ara-software/AraSim.git
This will download the github repo
Step 3:
We need to use make and load sourcing
- cd AraSim
- cvmfs
- make
wait and it should compile the code
Step 4:
We want to do a test run with 100 neutrinos to make sure that it does *actually* run
Try: - ./AraSim SETUP/setup.txt
This errored for me (probably you as well)
Switch from frequency domain to time domain in the setup.txt
- cd SETUP
- open setup.txt
- scroll to bottom
- Change SIMULATION_MODE = 1
- save
- cd ..
- ./AraSim SETUP/setup.txt
This should run quickly and now you have AraSim setup! |
|
1
|
Thu Mar 16 09:01:50 2017 |
Amy Connolly | Elog instructions | Other | Log into kingbee.mps.ohio-state.edu first, then log into radiorm.physics.ohio-state.edu.
From Keith Stewart 03/16/17: It appears that radiorm SSH from offsite is closed. So you will need to be on an OSU network physically or via VPN. fox is also blocked from offsite as well. Kingbee should still be available for now. If you want to use it as a jump host to get to radiorm without VPN. However, you will want to get comfortable with the VPN before it is a requirement.
Carl 03/16/17: I could log in even while using a hard line and plugged in directly to the network.
From Bryan Dunlap 12/16/17: I have set up the group permissions on the elog directory so you and your other designated people can edit files. I have configured sudo to allow you all to restart the elogd service. Once you have edited the file [/home/elog/elog.cfg I think], you can then type
sudo /sbin/service elogd restart
to restart the daemon so it re-reads the config. Sudo will prompt you for your password before it executes the command. |
|
12
|
Fri Aug 25 12:34:47 2017 |
Amy Connolly - posting stuff from Todd Thompson | How to run coffee | Other | One really useful thing in here is how to describe "Value Added" to visitors. |
| Attachment 1: guidelines.pdf
|
|
16
|
Thu Oct 26 08:44:58 2017 |
Brian Clark | Find Other Users on OSC | Other | Okay, so our group has two "project" spaces on OSC (the Ohio Supercomputer). The first is for Amy's group, and is a project workspace called "PAS0654". The second is the CCAPP Condo (literally, CCAPP has some pre-specified rental time, and hence "condo") on OSC, and this is project PCON0003.
When you are signed up for the supercomputer, one of two things happen:
- You will be given a username under the PAS0654 group, and in which case, your username will be something like osu****. Connolly home drive is /users/PAS0654/osu****. Beatty home drive is /users/PAS0174/osu****. CCAPP home drive is /users/PCON0003/pcon****.
- You will be given a username under the PCON0003 group, and in which case, your username will be something like cond****.
If you are given a osu**** username, you must make sure to be added to the CCAPP Condo so that you can use Ruby compute resources. It will not be automatic.
Some current group members, and their OSC usernames. In parenthesis is the project space they are found in.
Current Users
osu0673: Brian Clark (PAS0654)
cond0068: Jorge Torres-Espinosa (PCON0003)
osu8619: Keith McBride (PAS0174)
osu9348: Julie Rolla (PAS0654)
osu9979: Lauren Ennesser (PAS0654)
osu6665: Amy Connolly (PAS0654)
Past Users
osu0426: Oindree Banerjee (PAS0654)
osu0668: Brian Dailey (PAS0654)
osu8620: Jacob Gordon (PAS0174)
osu8386: Sam Stafford (ANITA analysis in /fs/scratch/osu8386) (PAS0174)
cond0091: Judge Rajasekera (PCON0003) |
|
19
|
Mon Mar 19 12:27:59 2018 |
Brian Clark | How To Do an ARA Monitoring Report | Other | So, ARA has five stations down in the ice that are taking data. Weekly, a member of the collaboration checks on the detectors to make sure that they are healthy.
This means things like making sure they are triggering at approximately the right rates, are taking cal pulsers, that the box isn't too hot, etc.
Here are some resources to get you started. Usual ara username and password apply in all cases.
Also, the page where all of the plots live is here: http://aware.wipac.wisc.edu/
Thanks, and good luck monitoring! Ask someone whose done it before when in doubt.
Brian |
|
40
|
Thu Jul 25 16:50:43 2019 |
Dustin Nguyen | Advice (not mine) on writing HEP stuff | Other | PDF of advice by Andy Buckley (U Glasgow) on writing a HEP thesis (and presumably HEP papers too) that was forwarded by John Beacom to the CCAPP mailing list a few months back. |
| Attachment 1: thesis-writing-gotchas.pdf
|
|
41
|
Fri Oct 11 13:43:35 2019 |
Amy | How to start a new undergrad hire | Other | If an undergrad has been working with the group on a volunteer basis, they will need that clarified, so that they have not up to then we working for pay without training. There is a form that they sign saying they have been working as a volunteer.
Below is an email that Pam sent in July 2019 outlining what they need. Other things to remember:
Undergrads receive emails from the department to remind them about orientation and scheduling prior to first day of hire, and other emails from ASC. They receive more information at orientation.
They will need to show ID.
If an undergrad (or any hire) does not waive retirement/OPERs within 30 days, they will have to have this deducted from their paycheck. This is another reason that orientation is important.
For the person hiring them:
1. Tell your admin (Lisa) asap who, when, etc you want to hire. ASAP is important because (Pam) will process over 75 undergraduates hires in the Fall and over 100 in the summer. Each takes 20 days on average (see below)
2. On the first day of employment – ask your new hire if they have completed their orientation with the ASC.
From: Hood, Pam
Sent: Tuesday, July 23, 2019 4:49 PM
To: 'physics-all@lists.osu.edu' <physics-all@lists.osu.edu>
Subject: Undergraduate Fall Hires
Hello all,
Fall is almost upon us and I am working on having positions ready to fill as well as posting on the OSU student job site and Federal Work study job boards. In order to plan my workflow, if you would let me know:
1. Approximately how many undergraduate student assistants you plan to hire and at what rate of pay. Our department’s current average rate of pay is $10.00 - $10.50/hour, however it does depend on the position. Range is $8.55 - $14.00/hour.
2. A brief summary of job duties and responsibilities (i.e. assisting in research for ______)
3. Request for posting OR
4. If you have specific student that has been a non-paid volunteer that you would like to hire, they need to sign a waiver prior to orientation. Please see attached for volunteer waiver.
5. Please indicate if your start date varies from August 20. And it may vary based on multiply factors i.e. signatures, workflow, etc.
6. All terminations for summer ungraduated student workers OR
7. If you are intending to continue employment but require a reduction in hours worked in order to comply with policy Please see attached policy for min/max hours.
I have attempted to answer all the FAQs that I noted in the “summer undergraduate hiring wave” in italics, however please let me know if you have questions. As always, it is ASC policy to not start any employee prior to orientation and it will take a minimum of 10 days from the time the contract is signed by both the student and the supervisor– please encourage your students to sign asap or it lengthens the process significantly.
I will do my best to facilitate this process for you. Students Buck ID# and email is extremely helpful. If you need the hire to start August 20, please send me the information via email by July 31, 2019. If you respond later or request a hire after July 31, I will estimate the hire date at the time of submittal.
Thanks,
Pam |
|
42
|
Tue Nov 5 16:22:16 2019 |
Keith McBride | NASA Proposal fellowships | Other | Here is a useful link for astroparticle grad students related proposals from NASA:
https://nspires.nasaprs.com/external/solicitations/summary.do?solId=%7BE16CD59F-29DD-06C0-8971-CE1A9C252FD4%7D&path=&method=init
Full email I received regarding this information was:
_______________________________________________________________________________________________________________________________________________________________________________________________________________________________________________
ROSES-19 Amendment adds a new program element to ROSES-2019: Future Investigators in NASA Earth and Space Science and Technology (FINESST), the ROSES Graduate Student Research Program Element, E.6.
Through FINESST, the Science Mission Directorate (SMD) solicits proposals from accredited U.S. universities and other eligible organizations for graduate student-designed and performed research projects that contribute to SMD's science, technology and exploration goals.
A Notice of Intent is not requested for E.6 FINESST. Proposals to FINESST are due by February 4, 2020.
Potential proposers who wish to participate in the optional pre-proposal teleconference December 2, 2019 from 1:00-2:30 p.m. Eastern Time may (no earlier than 30 minutes prior to the start time) call 1-888-324-3185 (U.S.-only Toll Free) or 1-630-395-0272 (U.S. Toll) and use Participant Passcode: 8018549. Restrictions may prevent the use of a toll-free number from a mobile or free-phone or from telephones outside the U.S. For U.S. TTY-equipped callers or other types of relay service no earlier than 30 minutes before the start of the teleconference, call 711 and provide the same conference call number/passcode. Email HQ-FINESST@mail.nasa.gov any teleconference agenda suggestions and questions by November 25, 2019. Afterwards, questions and responses, with identifying information removed, will be posted on the NSPIRES page for FINESST under "other documents".
FINESST awards research grants with a research mentor as the principal investigator and the listed graduate student listed as the "student participant". Unlike the extinct NASA Earth and Space Science Fellowships (NESSF), the Future Investigators (FIs) are not trainees or fellows. Students with existing NESSF awards seeking a third year of funding, may not submit proposals to FINESST. Instead, they may propose to NESSF20R. Subject to a period of performance restriction, some former NESSFs may be eligible to submit proposals to FINESST.
On or about November 1, 2019, this Amendment to the NASA Research Announcement "Research Opportunities in Space and Earth Sciences (ROSES) 2019" (NNH19ZDA001N) will be posted on the NASA research opportunity homepage at http://solicitation.nasaprs.com/ROSES2019 and will appear on the RSS feed at: https://science.nasa.gov/researchers/sara/grant-solicitations/roses-2019/
Questions concerning this program element may be directed to HQ-FINESST@mail.nasa.gov. |
|
5
|
Tue Apr 18 12:02:55 2017 |
Amy Connolly | How to ship to Pole | Hardware | Here is an old email thread about how to ship a station to Pole.
|
| Attachment 1: Shipping_stuff_to_Pole__a_short_how_to_from_you_would_be_nice_.pdf
|
| Attachment 2: ARA_12-13_UH_to_CHC_Packing_List_Box_2_of_3.pdf
|
| Attachment 3: ARA_12-13_UH_to_CHC_Packing_List_Box_1_of_3.pdf
|
| Attachment 4: ARA_12-13_UH_to_CHC_Packing_List_Box_3_of_3.pdf
|
| Attachment 5: IMG_4441.jpg
|  |
|
25
|
Wed Aug 1 11:37:10 2018 |
Andres Medina | Flux Ordering | Hardware | Bought 951 Non-Resin Soldering Flux. This is the preferred variety. This could be found on this website https://www.kester.com/products/product/951-soldering-flux.
The amount of Flux that was bought was 1 Gallon (Lasts quite some time). The price was $83.86 with approximate shipping of $43. This was done with a Pcard and a tax exempt form.
The website used to purchase this was https://www.alliedelec.com/kester-solder-63-0000-0951/70177935/ |
|
3
|
Wed Mar 22 18:01:23 2017 |
Brian Clark | Advice for Using the Ray Trace Correlator | Analysis | If you are trying to use the Ray Trace Correlator with AraRoot, you will probably encounter some issues as you go. Here is some advice that Carl Pfendner found, and Brian Clark compiled.
Please note that it is extremely important that your AntennaInfo.sqlite table in araROOT contain the ICRR versions of both Testbed and Station1. Testbed seems to have fallen out of practice of being included in the SQL table. Also, Station1 is the ICRR (earliest) version of A1, unlike the ATRI version which is logged as ARA01. This will cause seg faults in the intial setup of the timing and geometry arrays that seem unrelated to pure geometry files. If you get a seg-fault in the "setupSizes" function or the Detector call of the "setupPairs" function, checking your SQL file is a good idea. araROOT branch 3.13 has such a source table with Testbed and Station1 included.
Which combination of Makefile/Makefile.arch/StandardDefinitions.mk works can be machine specific (frustratingly). Sometimes the best StandardDefinitions.mk is found int he make_timing_arrays example.
Common Things to Check
1: Did you "make install" the Ray Trace Correlator after you made it?
2: Do you have the setup.txt file?
3: Do you have the "data" directory?
Common Errors
1: If the Ray Trace Correlator compiles, and you execute a binary, and get the following:
******** Begin Correlator ********, this one!
Pre-icemodel test
terminate called after throwing an instance of 'std::out_of_range'
what(): basic_string::substr
Aborted
Check to make sure have the "data" directory.
|
|
17
|
Mon Nov 20 08:31:48 2017 |
Brian Clark and Oindree Banerjee | Fit a Function in ROOT | Analysis | Sometimes you need to fit a function to a histogram in ROOT. Attached is code for how to do that in the simple case of a power law fit.
To run the example, you should type "root fitSnr.C" in the command line. The code will access the source histogram (hstrip1nsr.root, which is actually ANITA-2 satellite data). The result is stripe1snrfit.png. |
| Attachment 1: fitSnr.C
|
#include "TF1.h"
void fitSnr();
void fitSnr()
{
gStyle->SetLineWidth(4); //set some style parameters
TFile *stripe1file = new TFile("hstripe1snr.root"); //import the file containing the histogram to be fit
TH1D *hstripe1 = (TH1D*)stripe1file->Get("stripe1snr"); //get the histogram to be fit
TCanvas c1("c1","c1",1000,800); //make a canvas
hstripe1->Draw(""); //draw it
c1.SetLogy(); //set a log axis
//need to declare an equation
//I want to fit for two paramters, in the equation these are [0] and [1]
//so, you'll need to re-write the equation to whatever you're trying to fit for
//but ROOT wants the variables to find to be given as [0], [1], [2], etc.
char equation1[150]; //declare a container for the equation
sprintf(equation1,"([0]*(x^[1]))"); //declare the equation
TF1 *fit1 = new TF1("PowerFit",equation1,20,50); //create a function to fit with, with the range being 20 to 50
//now, we need to set the initial parameters of the fit
//fit->SetParameter(0,H->GetRMS()); //this should be a good starting place for a standard deviation like variable
//fit->SetParameter(1,H->GetMaximum()); //this should be a good starting place for amplitude like variable
fit1->SetParameter(0,60000.); //for our example, we will manually choose this
fit1->SetParameter(1,-3.);
hstripe1->Fit("PowerFit","R"); //actually do the fit;
fit1->Draw("same"); //draw the fit
//now, we want to print out some parameters to see how good the fit was
cout << "par0 " << fit1->GetParameter(0) << " par1 " << fit1->GetParameter(1) << endl;
cout<<"chisquare "<<fit1->GetChisquare()<<endl;
cout<<"Ndf "<<fit1->GetNDF()<<endl;
cout<<"reduced chisquare "<<double(fit1->GetChisquare())/double(fit1->GetNDF())<<endl;
cout<<" "<<endl;
c1.SaveAs("stripe1snrfit.png");
}
|
| Attachment 2: hstripe1snr.root
|
| Attachment 3: stripe1snrfit.png
| 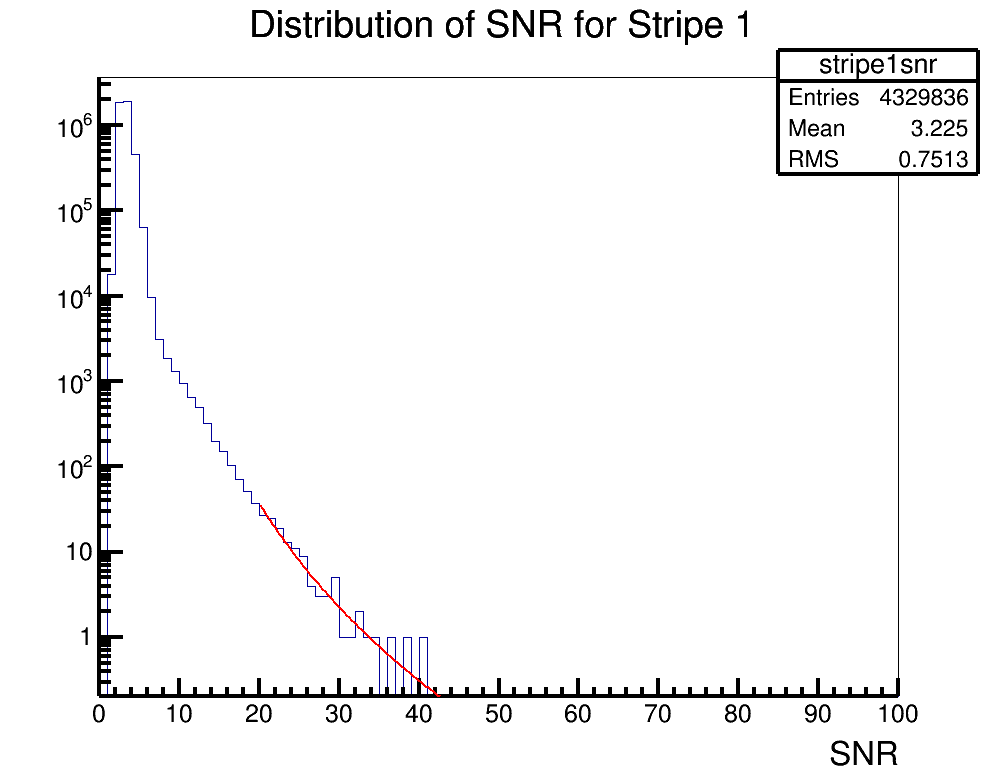 |
|
18
|
Tue Dec 12 17:38:36 2017 |
Brian Clark | Data Analysis in R from day w/ Brian Connolly | Analysis | On Nov 28 2017, Brian Connolly came and visited and taught us how to do basic data analysis in R.
He in particular showed us how to do a Linear Discriminant Analysis (LDA) and Principal Component Analysis (PCA).
Attached are three of the data files Carl Pfendner prepared for us to analyze (ARA data, including simulation, force triggers, and RF triggers).
Also attached is some R code that shows how to set up the LDA and the PCA and how to plot their result. You are meant to run each line of the code in r by hand (this is not a functioning R script I don't think).
Go here (https://www.r-project.org/) to learn how to download R. You will also probably have to download GGFortify. To do that, open an r session, and type "install.packages('ggfortify')". |
| Attachment 1: pca_and_lda.R
|
Notes
#First we need to read in the data
dataSim <- read.table("output.txt.Simulation",sep=" ",header=TRUE)
dataRF <- read.table("output.txt.RFTriggers",sep=" ",header=TRUE)
dataPulser <- read.table("output.txt.CalPulsers",sep=" ",header=TRUE)
#Now we combine them into one data object
data <- rbind(dataRF,dataSim)
#Now we actually have to specify that we
#want to use the first 10 columns
data <- data[,1:10]
#make a principal component analysis object
pca <- prcomp(data,center=TRUE,scale=TRUE,retx=TRUE)
#Load a plotting library
library(ggfortify)
#then can plot
autplot(prcomp(data))
#or, we can plot like this
#to get some color data points
labels<-c(rep(0,nrow(dataSim)),rep(1,nrow(dataRF)))
plot(pca$x[,1],pca$x[,2],col=labels+1)
#we can also do an LDA analysis
#we need to import the MASS library
library(MASS)
#and now we make the lda object
lda(data,grouping=labels)
#didn't write down any plotting unfortunately.
|
| Attachment 2: data.zip
|
|
22
|
Sun Apr 29 21:44:15 2018 |
Brian Clark | Access Deep ARA Station Data | Analysis | Quick C++ program for pulling waveforms out of deep ARA station data. If you are using AraRoot, you would put this inside your "analysis" directory and add it to your CMakeLists.txt. |
| Attachment 1: plot_deep_station_event.cxx
|
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
//// plot_event.cxx
//// plot deep station event
////
//// Apr 2018, clark.2668@osu.edu
////////////////////////////////////////////////////////////////////////////////
//Includes
#include <iostream>
//AraRoot Includes
#include "RawAtriStationEvent.h"
#include "UsefulAtriStationEvent.h"
#include "AraEventCalibrator.h"
//ROOT Includes
#include "TTree.h"
#include "TFile.h"
#include "TGraph.h"
#include "TCanvas.h"
using namespace std;
int main(int argc, char **argv)
{
//check to make sure they've given me a run and a pedestal
if(argc<2) {
std::cout << "Usage\n" << argv[0] << " <run file>\n";
std::cout << "e.g.\n" << argv[0] << " event1841.root\n";
return 0;
}
char pedFileName[200];
sprintf(pedFileName, "%s", argv[1]);
printf("------------------------------------------------------------------------\n");
printf("%s\n", argv[0]);
printf("runFileName %s\n", argv[1]);
printf("------------------------------------------------------------------------\n");
TFile *fp = TFile::Open(argv[2]);
if(!fp) {
std::cerr << "Can't open file\n";
return -1;
}
TTree *eventTree = (TTree*) fp->Get("eventTree");
if(!eventTree) {
std::cerr << "Can't find eventTree\n";
return -1;
}
RawAtriStationEvent *rawAtriEvPtr = 0; //empty pointer
eventTree->SetBranchAddress("event",&rawAtriEvPtr); //set the branch address
Int_t run_num; //run number of event
eventTree->SetBranchAddress("run", &run_num); //set the branch address
int numEntries=eventTree->GetEntries(); //get the number of events
int stationId=0;
eventTree->GetEntry(0);
stationId = rawAtriEvPtr->stationId; //assign the statio id number
AraEventCalibrator *calib = AraEventCalibrator::Instance(); //make a calibrator
for(int event=0;event<numEntries;event++) {
//for(int event=0;event<700;event++) {
eventTree->GetEntry(event); //get the event
int evt_num = rawAtriEvPtr->eventNumber; //check the event umber
if(rawAtriEvPtr->isCalpulserEvent()==0) continue; //bounce out if it's not a cal pulser
UsefulAtriStationEvent *realAtriEvPtr_fullcalib = new UsefulAtriStationEvent(rawAtriEvPtr, AraCalType::kLatestCalib); //make the event
TGraph *waveforms[16]={0};
for(int i=0; i<16; i++){
waveforms[i]=realAtriEvPtr_fullcalib->getGraphFromRFChan(i);
}
TCanvas *canvas = new TCanvas("","",1000,1000);
canvas->Divide(4,4);
for(int i=0; i<16; i++){
canvas->cd(i+1);
waveforms[i]->Draw("alp");
}
char title[200];
sprintf(title,"waveforms_run%d_event%d.pdf",stationId,run_num,evt_num);
canvas->SaveAs(title);
delete canvas;
}
}
|
|
27
|
Mon Oct 1 19:06:59 2018 |
Brian Clark | Code to Compute Effective Volumes in AraSim | Analysis | Here is some C++ code and an associated makefile to find effective volumes from AraSim output files.
It computes error bars on the effective volumes using the relevant AraSim function.
Compile like "make -f veff.mk"
Run like "./veff thrown_radius thrown_depth AraOut.1.root AraOut.2.root...." |
| Attachment 1: veff.cc
|
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include <cstdio>
#include <cstdlib>
#include <unistd.h>
#include <cstring>
#include <unistd.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TFile.h"
#include "TTree.h"
#include "TMath.h"
#include "Event.h"
#include "Detector.h"
#include "Report.h"
#include "Vector.h"
#include "counting.hh"
using namespace std;
//Get effective volumes with error bars
class EarthModel;
int main(int argc, char **argv)
{
gStyle->SetOptStat(111111);
gStyle->SetOptDate(1);
if(argc<4){
cout<<"Not enough arguments! Abort run."<<endl;
cout<<"Run like: ./veff volradius voldepth AraOut1.root AraOut2.rot ..."<<endl;
return -1;
}
double volradius = atof(argv[1]);
double voldepth = atof(argv[2]);
TChain *AraTree = new TChain("AraTree");
TChain *AraTree2 = new TChain("AraTree2");
for(int i=3; i<argc;i++){
AraTree->Add(string(argv[i]).c_str());
AraTree2->Add(string(argv[i]).c_str());
}
Report *report = 0;
Event *event=0;
AraTree2->SetBranchAddress("report",&report);
AraTree2->SetBranchAddress("event",&event);
int totnthrown = AraTree2->GetEntries();
cout << "Total number of events: " << totnthrown << endl;
int NBINS=10;
double eventsfound_binned[NBINS];
for(int i=0; i<NBINS; i++) eventsfound_binned[i]=0.;
double totweight=0;
for(int iEvt2=0; iEvt2<totnthrown; iEvt2++){
AraTree2->GetEntry(iEvt2);
if(report->stations[0].Global_Pass<=0) continue;
double weight = event->Nu_Interaction[0].weight;
if(weight > 1.0) continue;
totweight += weight;
int index_weights = Counting::findWeightBin(log10(weight));
if(index_weights<NBINS) eventsfound_binned[index_weights]++;
}
double error_minus=0.;
double error_plus=0.;
Counting::findErrorOnSumWeights(eventsfound_binned,error_plus,error_minus);
double vtot = TMath::Pi() * double(volradius) * double(volradius) * double(voldepth) / 1.e9; //answer in km^3
double veff = vtot * totweight / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
double veff_p = vtot * (error_plus) / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
double veff_m = vtot * (error_minus) / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
printf("volthrown: %.6f \n totweight: %.6f + %.6f - %.6f \n Veff: %.6f + %.6f - %.6f \n",
vtot,
totweight, error_plus, error_minus,
veff, veff_p, veff_m
);
return 0;
}
|
| Attachment 2: veff.mk
|
#############################################################################
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o veff.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc veff.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = veff
all : $(PROGRAMS)
veff : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
|
28
|
Fri Oct 26 18:08:43 2018 |
Jorge Torres | Analyzing effective volumes | Analysis | Attaching some scripts that help processing the effective volumes. This is an extension of what Brian Clark did in a previous post (http://radiorm.physics.ohio-state.edu/elog/How-To/27)
There are 4 files attached:
- veff_aeff2.C and veff_aeff2.mk. veff_aeff2.C produces Veff_des$1.txt ($1 can be A or B or C). This file contains the following columns: energy, veff, veff_error, veff1 (PA), veff2 (LPDA), veff3 (bicone), respectively. However, the energies are not sorted.
-veff.sh: this bash executable runs veff_aeff2.C for all (that's what the "*" in the executable is for) the root output files, for a given design (A, B, C). You need to modify the location of your output files, though. Run like "./veff.sh A", which will execute veff_aeff2.C and produce the veff text files. Do the same for B or C.
-make_plot.py: takes Veff_des$1.txt, sorts energies out, plots the effective volumes vs. energies, and produces a csv file containing the veffs (just for the sake of copying and pastting on the spreadsheets). Run like "pyhton make_plot.py".
|
| Attachment 1: veff.sh
|
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_16.5.txt.run* &
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_17.txt.run* &
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_17.5.txt.run* &
nohup ./veff_aeff2 5000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_18.txt.run* &
nohup ./veff_aeff2 5000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_18.5.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_19.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_19.5.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_20.txt.run* &
|
| Attachment 2: veff_aeff2.C
|
//////////////////////////////////////
//To calculate various veff and aeff
//Run as ./veff_aeff2 $RADIUS $DEPTH $FILE
//For example ./veff_aeff2 8000 3000 /data/user/ypan/bin/AraSim/trunk/outputs/AraOut.setup_single_station_2_energy_20.run0.root
//
/////////////////////////////////////
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TH1.h"
#include "TF1.h"
#include "TF2.h"
#include "TFile.h"
#include "TRandom.h"
#include "TRandom2.h"
#include "TRandom3.h"
#include "TTree.h"
#include "TLegend.h"
#include "TLine.h"
#include "TROOT.h"
#include "TPostScript.h"
#include "TCanvas.h"
#include "TH2F.h"
#include "TText.h"
#include "TProfile.h"
#include "TGraphErrors.h"
#include "TStyle.h"
#include "TMath.h"
#include <unistd.h>
#include "TVector3.h"
#include "TRotation.h"
#include "TSpline.h"
//#include "TObject.h"
#include "Tools.h"
#include "Constants.h"
#include "Vector.h"
#include "Position.h"
#include "EarthModel.h"
#include "IceModel.h"
#include "Efficiencies.h"
#include "Spectra.h"
#include "Event.h"
#include "Trigger.h"
#include "Detector.h"
#include "Settings.h"
#include "counting.hh"
#include "Primaries.h"
#include "signal.hh"
#include "secondaries.hh"
#include "Ray.h"
#include "RaySolver.h"
#include "Report.h"
using namespace std;
class EarthModel;
int main(int argc, char **argv){
//string readfile;
//readfile = string(argv[1]);
//readfile = "/data/user/ypan/bin/AraSim/branches/new_geom/outputs/AraOut20.0.root";
int ifile = 0;
double totweightsq;
double totweight;
int totnthrown;
int typetotnthrown[12];
double tottrigeff;
double sigma[12];
double typetotweight[12];
double typetotweightsq[12];
double totsigmaweight;
double totsigmaweightsq;
double volradius;
double voldepth;
const int nstrings = 9;
const int nantennas = 1;
double veff1 = 0.0;
double veff2 = 0.0;
double veff3 = 0.0;
double weight1 = 0.0;
double weight2 = 0.0;
double weight3 = 0.0;
double veffT[6], vefferrT[6], aeffT[6], aefferrT[6];
double veffF[3], vefferrF[3], aeffF[3], aefferrF[3];
double veffNu[2], vefferrNu[2], aeffNu[2], aefferrNu[2];
double veff, vefferr, aeff, aefferr, aeff2;
double pnu;
Detector *detector = 0;
//Settings *settings = 0;
//IceModel *icemodel = 0;
Event *event = 0;
Report *report = 0;
cout<<"construct detector"<<endl;
//TFile *AraFile=new TFile(readfile.c_str());
//TFile *AraFile=new TFile((outputdir+"/AraOut.root").c_str());
TChain *AraTree = new TChain("AraTree");
TChain *AraTree2 = new TChain("AraTree2");
TChain *eventTree = new TChain("eventTree");
//AraTree->SetBranchAddress("detector",&detector);
//AraTree->SetBranchAddress("settings",&settings);
//AraTree->SetBranchAddress("icemodel",&icemodel);
cout << "trees set" << endl;
for(ifile = 3; ifile < (argc - 1); ifile++){
AraTree->Add(string(argv[ifile + 1]).c_str());
AraTree2->Add(string(argv[ifile + 1]).c_str());
eventTree->Add(string(argv[ifile + 1]).c_str());
}
AraTree2->SetBranchAddress("event",&event);
AraTree2->SetBranchAddress("report",&report);
cout<<"branch detector"<<endl;
for(int i=0; i<12; i++) {
typetotweight[i] = 0.0;
typetotweightsq[i] = 0.0;
typetotnthrown[i] = 0;
}
totweightsq = 0.0;
totweight = 0.0;
totsigmaweight = 0.0;
totsigmaweightsq = 0.0;
totnthrown = AraTree2->GetEntries();
cout << "Total number of events: " << totnthrown << endl;
//totnthrown = settings->NNU;
//volradius = settings->POSNU_RADIUS;
volradius = atof(argv[1]);
voldepth = atof(argv[2]);
AraTree2->GetEntry(0);
pnu = event->pnu;
cout << "Energy " << pnu << endl;
for(int iEvt2=0; iEvt2<totnthrown; iEvt2++) {
AraTree2->GetEntry(iEvt2);
double sigm = event->Nu_Interaction[0].sigma;
int iflavor = (event->nuflavorint)-1;
int inu = event->nu_nubar;
int icurr = event->Nu_Interaction[0].currentint;
sigma[inu+2*icurr+4*iflavor] = sigm;
typetotnthrown[inu+2*icurr+4*iflavor]++;
if( (iEvt2 % 10000 ) == 0 ) cout << "*";
if(report->stations[0].Global_Pass<=0) continue;
double weight = event->Nu_Interaction[0].weight;
if(weight > 1.0){
cout << weight << "; " << iEvt2 << endl;
continue;
}
// cout << weight << endl;
totweightsq += pow(weight,2);
totweight += weight;
typetotweight[inu+2*icurr+4*iflavor] += weight;
typetotweightsq[inu+2*icurr+4*iflavor] += pow(weight,2);
totsigmaweight += weight*sigm;
totsigmaweightsq += pow(weight*sigm,2);
int trig1 = 0;
int trig2 = 0;
int trig3 = 0;
for (int i = 0; i < nstrings; i++){
if (i == 0 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig1++;
if (i > 0 && i < 5 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig2++;
if (i > 4 && i < 9 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig3++;
}
if ( trig1 > 0)//phase array
weight1 += event->Nu_Interaction[0].weight;
if ( trig2 > 1)//lpda
weight2 += event->Nu_Interaction[0].weight;
if (trig3 > 3)//bicone
weight3 += event->Nu_Interaction[0].weight;
}
tottrigeff = totweight / double(totnthrown);
double nnucleon = 5.54e29;
double vtot = PI * double(volradius) * double(volradius) * double(voldepth) / 1e9;
veff = vtot * totweight / double(totnthrown) * 4.0 * PI;
//vefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totweightsq)/totweight));
vefferr = sqrt(totweightsq) / totweight * veff;
aeff = vtot * (1e3) * nnucleon * totsigmaweight / double(totnthrown);
//aefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totsigmaweightsq)/totsigmaweight));
//aefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totweightsq)/totweight));
aefferr = sqrt(totweightsq) / totweight * aeff;
double sigmaave = 0.0;
for(int iflavor=0; iflavor<3; iflavor++) {
double flavorweight = 0.0;
double flavorweightsq = 0.0;
double flavorsigmaave = 0.0;
int flavortotthrown = 0;
double temptotweightnu[2] = {0};
double tempsignu[2] = {0};
double temptotweight = 0.0;
for(int inu=0; inu<2; inu++) {
double tempsig = 0.0;
double tempweight = 0.0;
for(int icurr=0; icurr<2; icurr++) {
tempsig += sigma[inu+2*icurr+4*iflavor];
tempsignu[inu] += sigma[inu+2*icurr+4*iflavor];
tempweight += typetotweight[inu+2*icurr+4*iflavor];
flavorweight += typetotweight[inu+2*icurr+4*iflavor];
flavorweightsq += typetotweightsq[inu+2*icurr+4*iflavor];
temptotweight += typetotweight[inu+2*icurr+4*iflavor];
temptotweightnu[inu] += typetotweight[inu+2*icurr+4*iflavor];
flavortotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
//printf("Temp Sigma: "); cout << tempsig << "\n";
sigmaave += tempsig*(tempweight/totweight);
}
flavorsigmaave += tempsignu[0]*(temptotweightnu[0]/temptotweight)+tempsignu[1]*(temptotweightnu[1]/temptotweight);
veffF[iflavor] = vtot*flavorweight/double(flavortotthrown);
vefferrF[iflavor] = sqrt(flavorweightsq)/flavorweight;
//printf("Volume: %.9f*%.9f/%.9f \n",vtot,flavorweight,double(totnthrown));
aeffF[iflavor] = veffF[iflavor]*(1e3)*nnucleon*flavorsigmaave;
aefferrF[iflavor] = sqrt(flavorweightsq)/flavorweight;
}
for(int inu=0; inu<2; inu++) {
double tempsig = 0.0;
double tempweight = 0.0;
double tempweightsq = 0.0;
int nutotthrown = 0;
for(int iflavor=0; iflavor<3; iflavor++) {
for(int icurr=0; icurr<2; icurr++) {
tempweight += typetotweight[inu+2*icurr+4*iflavor];
tempweightsq += typetotweightsq[inu+2*icurr+4*iflavor];
nutotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
}
tempsig += sigma[inu+2*0+4*0];
tempsig += sigma[inu+2*1+4*0];
veffNu[inu] = vtot*tempweight/double(nutotthrown);
vefferrNu[inu] = sqrt(tempweightsq)/tempweight;
aeffNu[inu] = veffNu[inu]*(1e3)*nnucleon*tempsig;
aefferrNu[inu] = sqrt(tempweightsq)/tempweight;
}
double totalveff = 0.0;
double totalaeff = 0.0;
for(int inu=0; inu<2; inu++) {
for(int iflavor=0; iflavor<3; iflavor++) {
int typetotthrown = 0;
for(int icurr=0; icurr<2; icurr++) {
typetotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
totalveff += veffT[iflavor+3*inu]*(double(typetotthrown)/double(totnthrown));
totalaeff += aeffT[iflavor+3*inu]*(double(typetotthrown)/double(totnthrown));
}
}
aeff2 = veff*(1e3)*nnucleon*sigmaave;
aeff = aeff2;
veff1 = weight1 / totnthrown * vtot * 4.0 * PI;
veff2 = weight2 / totnthrown * vtot * 4.0 * PI;
veff3 = weight3 / totnthrown * vtot * 4.0 * PI;
printf("\nvolthrown: %.6f; totweight: %.6f; Veff: %.6f +- %.6f\n", vtot, totweight, veff, vefferr);
printf("veff1: %.3f; veff2: %.3f; veff3: %.3f\n", veff1, veff2, veff3);
//string des = string(argv[4]);
char buf[100];
std::ostringstream stringStream;
stringStream << string(argv[3]);
std::string copyOfStr = stringStream.str();
snprintf(buf, sizeof(buf), "Veff_des%s.txt", copyOfStr.c_str());
FILE *fout = fopen(buf, "a+");
fprintf(fout, "%e, %.6f, %.6f, %.3f, %.3f, %.3f \n", pnu, veff, vefferr, veff1, veff2, veff3);
fclose(fout);
return 0;
}
|
| Attachment 3: veff_aeff2.mk
|
#############################################################################
## Makefile -- New Version of my Makefile that works on both linux
## and mac os x
## Ryan Nichol <rjn@hep.ucl.ac.uk>
##############################################################################
##############################################################################
##############################################################################
##
##This file was copied from M.readGeom and altered for my use 14 May 2014
##Khalida Hendricks.
##
##Modified by Brian Clark for use on CosTheta_NuTraject on 20 February 2015
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##
##############################################################################
##############################################################################
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o veff_aeff2.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc veff_aeff2.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = veff_aeff2
all : $(PROGRAMS)
veff_aeff2 : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
| Attachment 4: make_plot.py
|
# -*- coding: utf-8 -*-
import numpy as np
import sys
import matplotlib.pyplot as plt
from pylab import setp
from matplotlib.pyplot import rcParams
import csv
import pandas as pd
rcParams['mathtext.default'] = 'regular'
def read_file(finame):
fi = open(finame, 'r')
rdr = csv.reader(fi, delimiter=',', skipinitialspace=True)
table = []
for row in rdr:
# print(row)
energy = float(row[0])
veff = float(row[1])
veff_err = float(row[2])
veff1 = float(row[3])
veff2 = float(row[4])
veff3 = float(row[5])
row = {'energy':energy, 'veff':veff, 'veff_err':veff_err, 'veff1':veff1, 'veff2':veff2, 'veff3':veff3}
table.append(row)
df=pd.DataFrame(table)
df_ordered=df.sort_values('energy',ascending=True)
# print(df_ordered)
return df_ordered
def beautify_veff(this_ax):
sizer=20
xlow = 1.e16 #the lower x limit
xup = 2.e20 #the uppper x limit
ylow =1e-3 #the lower x limit
yup = 6.e1 #the uppper x limit
this_ax.set_xlabel('Energy [eV]',size=sizer) #give it a title
this_ax.set_ylabel('[V$\Omega]_{eff}$ [km$^3$sr]',size=sizer)
this_ax.set_yscale('log')
this_ax.set_xscale('log')
this_ax.tick_params(labelsize=sizer)
this_ax.set_xlim([xlow,xup]) #set the x limits of the plot
this_ax.set_ylim([ylow,yup]) #set the y limits of the plot
this_ax.grid()
this_legend = this_ax.legend(loc='upper left')
setp(this_legend.get_texts(), fontsize=17)
setp(this_legend.get_title(), fontsize=17)
def main():
"""
arasim_energies = np.array([3.16e+16, 1e+17, 3.16e+17, 1e+18, 3.16e+18, 1e+19, 3.16e+19, 1e+20])
arasim_energies2 = np.array([3.16e+16, 1e+17, 3.16e+17, 1e+18, 1e+19, 3.16e+19, 1e+20])
#arasim_desA_veff = np.array([0.080894,0.290695,0.943223,2.388708,4.070498,6.824112,10.506490,13.969418])
arasim_desA_veff_s = np.array([0.067384,0.289591,0.996509,2.464464,4.945600,8.735506,13.357300,18.751915])
# arasim_desA_veff_ice = np.array([0.066291,0.303620,0.927647,2.427554,4.962093,8.465895,13.425852,18.706528])
arasim_desA_error = np.array([0.008367,0.017401,0.032222,0.084223,0.119240,0.221266,0.272460,0.320456])
# arasim_desA_error_ice = np.array([0.008345,0.017748,0.031295,0.083747,0.119370,0.217717,0.273075,0.319872])
# arasim_desB_veff = np.array([0.124111,0.417102,1.310555,3.648494,4.070E+0,11.675189,18.393961,13.688909])
arasim_desB_veff_s = np.array([0.064937,0.355747,1.289947,3.821705,8.002805,15.352981,25.391282,18.009545])
# arasim_desB_veff_ice = np.array([0.080070,0.340927,1.369081,3.938550,8.211407,15.190858,25.066541,18.021033])
arasim_desB_error = np.array([0.008268,0.019317,0.036926,0.105445,0.152488,0.296617,0.377997,0.314314])
# arasim_desB_error_ice = np.array([0.009220,0.019144,0.037966,0.107189,0.154430,0.293728,0.375827,0.314244])
arasim_desA_veff1_sm = np.array([0.054,0.231,0.868,2.239,4.586,8.339,12.910,18.301])
arasim_desA_veff2_sm = np.array([0.009,0.036,0.098,0.323,0.705,1.167,2.169,3.130])
arasim_desA_veff3_sm = np.array([0.011,0.074,0.193,0.664,1.208,2.347,3.689,4.814])
arasim_desA_veff1 = np.array([0.053,0.282,1.108,3.462,7.486,14.613,24.682,17.557])
arasim_desA_veff2 = np.array([0.006,0.040,0.121,0.322,0.636,1.308,1.961,2.708])
arasim_desA_veff3 = np.array([0.006,0.065,0.188,0.638,1.187,2.270,3.301,4.322])
"""
veff_A=read_file("Veff_desA.txt")
veff_B=read_file("Veff_desB.txt")
print("desA is \n", veff_A)
print("desB is \n", veff_B)
dfA = veff_A[['veff','veff_err']]
dfB = veff_B[['veff','veff_err']]
dfA.to_csv('veffA.csv', sep='\t',index=False)
dfB.to_csv('veffB.csv', sep='\t',index=False)
fig = plt.figure(figsize=(11,8.5))
ax1 = fig.add_subplot(1,1,1)
ax1.plot(veff_A['energy'], veff_A['veff'],'bs-',label='Strawman',markersize=8,linewidth=2)
ax1.plot(veff_B['energy'], veff_B['veff'],'gs-',label='Punch @100 m',markersize=8,linewidth=2)
ax1.fill_between(veff_A['energy'], veff_A['veff']-veff_A['veff_err'], veff_A['veff']+veff_A['veff_err'], alpha=0.2, color='red')
ax1.fill_between(veff_B['energy'], veff_B['veff']-veff_B['veff_err'], veff_B['veff']+veff_B['veff_err'], alpha=0.2, color='red')
beautify_veff(ax1)
ax1.set_title("Punch vs Strawman, noise + signal",fontsize=20)
fig.savefig("desAB.png",edgecolor='none',bbox_inches="tight") #save the figure
fig2 = plt.figure(figsize=(11,8.5))
ax2 = fig2.add_subplot(1,1,1)
#Triggers plot
ax2.plot(veff_B['energy'], veff_B['veff1'],'g^-',label='Phased array (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff1'],'gs-',label='Phased array (Strawman)',markersize=8,linewidth=2)
ax2.plot(veff_B['energy'], veff_B['veff2'],'b^-',label='LPDAs (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff2'],'bs-',label='LPDAs (Strawman)',markersize=8,linewidth=2)
ax2.plot(veff_B['energy'], veff_B['veff3'],'y^-',label='Bicones (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff3'],'ys-',label='Bicones (Strawman)',markersize=8,linewidth=2)
ax2.set_title("Triggers contribution, noise + signal",fontsize=20)
beautify_veff(ax2)
fig2.savefig("desAB_triggers.png",edgecolor='none',bbox_inches="tight") #save the figure
main()
|
|
30
|
Tue Nov 27 09:43:37 2018 |
Brian Clark | Plot Two TH2Ds with Different Color Palettes | Analysis | Say you want to plot two TH2Ds with on the same pad but different color paletettes?
This is possible, but requires a touch of fancy ROOT-ing. Actually, it's not that much, it just takes a lot of time to figure it out. So here it is.
Fair warning, the order of all the gPad->Modified's etc seems very important to no seg fault.
In include the main (demo.cc) and the Makefile. |
| Attachment 1: demo.cc
|
///////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
/////// demo.cxx
////// Nov 2018, Brian Clark
////// Demonstrate how to draw 2 TH2D's with two different color palettes (needs ROOT6)
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
//C++
#include <iostream>
//ROOT Includes
#include "TH2D.h"
#include "TCanvas.h"
#include "TStyle.h"
#include "TExec.h"
#include "TColor.h"
#include "TPaletteAxis.h"
#include "TRandom3.h"
using namespace std;
int main(int argc, char **argv)
{
gStyle->SetOptStat(0);
TH2D *h1 = new TH2D("h1","h1",100,-10,10,100,-10,10);
TH2D *h2 = new TH2D("h2","h2",100,-10,10,100,-10,10);
TRandom3 *R = new TRandom3(time(0));
int count=0;
while(count<10000000){
h1->Fill(R->Gaus(-4.,1.),R->Gaus(-4.,1.));
count++;
}
TRandom3 *R2 = new TRandom3(time(0)+100);
count=0;
while(count<10000000){
h2->Fill(R2->Gaus(4.,1.),R2->Gaus(4.,1.));
count++;
}
TCanvas *c = new TCanvas("c","c",1100,850);
h1->Draw("colz");
TExec *ex1 = new TExec("ex1","gStyle->SetPalette(kValentine);");
ex1->Draw();
h1->Draw("colz same");
gPad->SetLogz();
gPad->Modified();
gPad->Update();
TPaletteAxis *palette1 = (TPaletteAxis*)h1->GetListOfFunctions()->FindObject("palette");
palette1->SetX1NDC(0.7);
palette1->SetX2NDC(0.75);
palette1->SetY1NDC(0.1);
palette1->SetY2NDC(0.95);
gPad->SetRightMargin(0.3);
gPad->SetTopMargin(0.05);
h2->Draw("same colz");
TExec *ex2 = new TExec("ex2","gStyle->SetPalette(kCopper);"); //TColor::InvertPalette();
ex2->Draw();
h2->Draw("same colz");
gPad->SetLogz();
gPad->Modified();
gPad->Update();
TPaletteAxis *palette2 = (TPaletteAxis*)h2->GetListOfFunctions()->FindObject("palette");
palette2->SetX1NDC(0.85);
palette2->SetX2NDC(0.90);
palette2->SetY1NDC(0.1);
palette2->SetY2NDC(0.95);
h1->SetTitle("");
h1->GetXaxis()->SetTitle("X-Value");
h1->GetYaxis()->SetTitle("Y-Value");
h1->GetZaxis()->SetTitle("First Histogram Events");
h2->GetZaxis()->SetTitle("Second Histogram Events");
h1->GetYaxis()->SetTitleSize(0.05);
h1->GetXaxis()->SetTitleSize(0.05);
h1->GetYaxis()->SetLabelSize(0.045);
h1->GetXaxis()->SetLabelSize(0.045);
h1->GetZaxis()->SetTitleSize(0.045);
h2->GetZaxis()->SetTitleSize(0.045);
h1->GetZaxis()->SetLabelSize(0.04);
h2->GetZaxis()->SetLabelSize(0.04);
h1->GetYaxis()->SetTitleOffset(1);
h1->GetZaxis()->SetTitleOffset(1);
h2->GetZaxis()->SetTitleOffset(1);
c->SetLogz();
c->SaveAs("test.png");
}
|
| Attachment 2: Makefile
|
LDFLAGS=-L${ARA_UTIL_INSTALL_DIR}/lib -L${shell root-config --libdir}
CXXFLAGS=-I${ARA_UTIL_INSTALL_DIR}/include -I${shell root-config --incdir}
LDLIBS += $(shell $(ROOTSYS)/bin/root-config --libs)
|
| Attachment 3: test.png
| 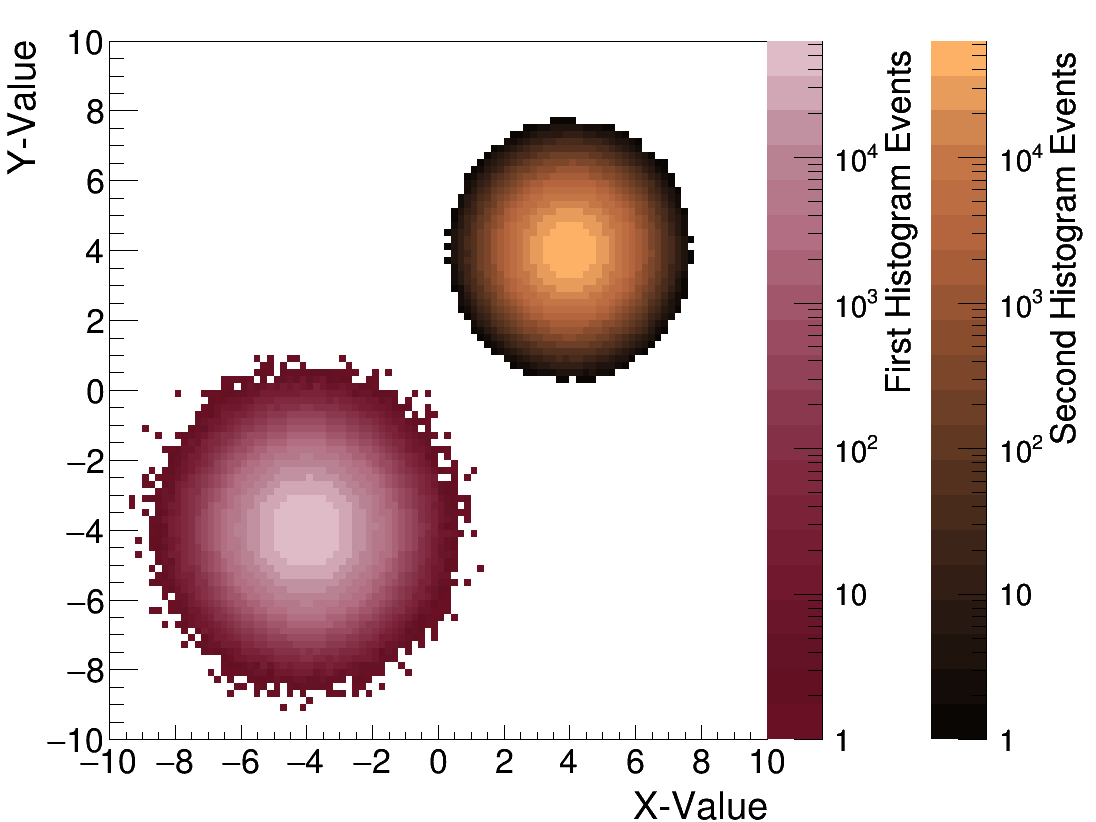 |
|
36
|
Mon Mar 4 14:11:07 2019 |
Jorge Torres | Submitting arrays of jobs on OSC | Analysis | PBS has the option of having arrays of jobs in case you want to easily submit multiple similar jobs. The only difference in them is the array index, which you can use in your PBS script to run each task with a different set of input arguments, or any other operation that requires a unique index.
You need to add the following lines to your submitter file:
#PBS -t array_min-array_max%increment
where "array_min/array_max" are integers that set lower and upper limit, respectively, of your "job loop" and increment lets you set the number of jobs that you want to submit simultaneously. For example:
submits an array with 100 jobs in it, but the system will only ever have 5 running at one time.
Here's an example of a script that submits to Pitzer a job array (from 2011-3000 in batches of 40) of a script named "make_fits_noise" that uses one core only. Make sure you use "#PBS -m n", otherwise you'll get tons of emails notyfing you about your jobs.
To delete the whole array, use "qdel JOB_ID[ ]". To delete one single instance, use "qdel JOB_ID[ARRAY_ID]".
More info: https://arc-ts.umich.edu/software/torque/job-arrays/
|
|
2
|
Thu Mar 16 10:39:15 2017 |
Amy Connolly | How Do I Connect to the ASC VPN Using Cisco and Duo? | | For Mac and Windows:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=14542
For Linux, in case some of your students need it:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=17908
From Sam 01/25/17: It doesn't work from my Ubuntu 14 machine. My VPN setup in 14 does not have the "Software Token Authentication" option on the screen as shown in the instructions. It fails on connection attempt.
The instructions specify Ubuntu 16; perhaps there is a way to make it work on 14, but I don't know what it is.
|
|
Draft
|
Fri Jul 28 17:57:06 2017 |
Brian Clark and Ian Best | | | |
|