| ID |
Date |
Author |
Type |
Category |
Subject |
Project |
|
29
|
Tue Feb 27 23:13:15 2018 |
Amy Connolly | Other | Simulation | Plots for QC sites | | Hi All,
We need to revive the icemcQC and AraSimQC pages, and I thought that in the meantime we could keep a list and/or associated code for plots we'd like to see go in there.
Here are a couple plots I just made code attached is forstevenanddave.cc
event weight (attenuation factor) vs. angle that neutrino makes wrt up when standing under the anita balloon
event weight (attenuation factor) vs. angle that rf makes wrt up from an observer on the anita balloon
BC: Adding old Stephen Hoover plots from ANITA elog: https://www.phys.hawaii.edu/elog/anita_notes/32
|
| Attachment 1: forstevenanddave.cc
|
#include <iostream>
#include <fstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
//#include <cmath>
#include "TH1.h"
#include "TFile.h"
#include "TLine.h"
#include "TTree.h"
#include "TROOT.h"
#include "TPostScript.h"
#include "TCanvas.h"
#include "TH2F.h"
#include "TText.h"
#include "TProfile.h"
#include "TGraphErrors.h"
#include "TRandom.h"
#include "TRandom2.h"
#include "TRandom3.h"
#include "TStyle.h"
#include "TVector3.h"
using namespace std;
TStyle* RootStyle();
TStyle *color=RootStyle();
double PI=3.1415926;
double R_EARTH=6.36E6;
TRandom3 *Rand3=new TRandom3();
int main(int argc, char *argv[]) {
gStyle=color;
gStyle->SetPalette(1);
TFile *f = new TFile("/Users/amyc/icemc/outputs/icefinal.root");
TTree *t1 = (TTree*)f->Get("passing_events");
double logweight,theta_in,costhetanu,cosalpha,phi_in;
double theta_rf_atbn;
double e_component,h_component;
double weight;
t1->SetBranchAddress("logweight",&logweight);
t1->SetBranchAddress("weight",&weight);
// t1->SetBranchAddress("theta_in",&theta_in);
//t1->SetBranchAddress("phi_in",&phi_in);
//t1->SetBranchAddress("costhetanu",&costhetanu);
t1->SetBranchAddress("cosalpha",&cosalpha);
t1->SetBranchAddress("theta_rf_atbn",&theta_rf_atbn);
t1->SetBranchAddress("e_component",&e_component);
t1->SetBranchAddress("h_component",&h_component);
TH2D *h2_weight_thetarf=new TH2D("h2_thetarf","h2_thetarf",100,50.,100.,100,-10.,1.);
TH2D *h2_weight_alpha=new TH2D("h2_alpha","h2_alpha",100,90.0,130.,100,-10.,1.);
TH1D *h2_ratio=new TH1D("h2_ratio","h2_ratio",100,-20.0,20.);
for (int i=0;i<t1->GetEntries();i++) {
t1->GetEvent(i);
h2_weight_thetarf->Fill(theta_rf_atbn*180./PI,logweight);
h2_weight_alpha->Fill(acos(cosalpha)*180./PI,logweight);
h2_ratio->Fill(e_component/h_component,1E5*weight);
cout << "weight, theta are " << logweight << "\t" << theta_rf_atbn << "\n";
}
TCanvas *c1=new TCanvas("c1","c1",800,800);
c1->Divide(1,2);
h2_weight_thetarf->SetXTitle("#theta_{rf at bn}");
h2_weight_thetarf->SetYTitle("Weight");
c1->cd(1);
h2_weight_thetarf->SetMarkerSize(0.5);
h2_weight_thetarf->Draw();
c1->cd(2);
h2_weight_alpha->SetXTitle("#theta_{#nu wrt up}");
h2_weight_alpha->SetYTitle("Weight");
h2_weight_alpha->Draw();
c1->cd(2);
h2_weight_alpha->SetXTitle("#alpha_{rf at bn}");
h2_weight_alpha->SetYTitle("Weight");
h2_weight_alpha->Draw();
c1->Print("stevenanddave.eps");
return 0;
}
TStyle* RootStyle() {
// const char* modified = "Borrowed and adapted from paus et al";
TStyle *RootStyle = new TStyle("Root-Style","The Perfect Style for Plots ;-)");
#ifdef __CINT__
TStyle *GloStyle;
GloStyle = gStyle; // save the global style reference
gStyle = RootStyle;
#endif
// otherwise you need to call TROOT::SetStyle("Root-Style")
// Paper size
RootStyle->SetPaperSize(TStyle::kUSLetter);
// Canvas
RootStyle->SetCanvasColor (0);
RootStyle->SetCanvasBorderSize(10);
RootStyle->SetCanvasBorderMode(0);
RootStyle->SetCanvasDefH (600);
RootStyle->SetCanvasDefW (600);
RootStyle->SetCanvasDefX (10);
RootStyle->SetCanvasDefY (10);
// Pads
RootStyle->SetPadColor (0);
RootStyle->SetPadBorderSize (10);
RootStyle->SetPadBorderMode (0);
// RootStyle->SetPadBottomMargin(0.13);
RootStyle->SetPadBottomMargin(0.16);
RootStyle->SetPadTopMargin (0.08);
RootStyle->SetPadLeftMargin (0.15);
RootStyle->SetPadRightMargin (0.15);
RootStyle->SetPadGridX (0);
RootStyle->SetPadGridY (0);
RootStyle->SetPadTickX (1);
RootStyle->SetPadTickY (1);
// Frames
RootStyle->SetFrameFillStyle ( 0);
RootStyle->SetFrameFillColor ( 0);
RootStyle->SetFrameLineColor ( 1);
RootStyle->SetFrameLineStyle ( 0);
RootStyle->SetFrameLineWidth ( 2);
RootStyle->SetFrameBorderSize(10);
RootStyle->SetFrameBorderMode( 0);
// Histograms
RootStyle->SetHistFillColor(0);
RootStyle->SetHistFillStyle(1);
RootStyle->SetHistLineColor(1);
RootStyle->SetHistLineStyle(0);
RootStyle->SetHistLineWidth(2);
// Functions
RootStyle->SetFuncColor(1);
RootStyle->SetFuncStyle(0);
RootStyle->SetFuncWidth(1);
//Legends
RootStyle->SetStatBorderSize(2);
RootStyle->SetStatFont (42);
// RootStyle->SetOptStat (111111);
RootStyle->SetOptStat (0);
RootStyle->SetStatColor (0);
RootStyle->SetStatX (0.93);
RootStyle->SetStatY (0.90);
RootStyle->SetStatFontSize (0.07);
// RootStyle->SetStatW (0.2);
// RootStyle->SetStatH (0.15);
// Labels, Ticks, and Titles
RootStyle->SetTickLength ( 0.015,"X");
RootStyle->SetTitleSize ( 0.055,"X");
RootStyle->SetTitleOffset( 1.00,"X");
RootStyle->SetTitleBorderSize(0);
// RootStyle->SetTitleFontSize((float)3.);
RootStyle->SetLabelOffset( 0.015,"X");
RootStyle->SetLabelSize ( 0.050,"X");
RootStyle->SetLabelFont ( 42 ,"X");
RootStyle->SetTickLength ( 0.015,"Y");
RootStyle->SetTitleSize ( 0.055,"Y");
RootStyle->SetTitleOffset( 1.300,"Y");
RootStyle->SetLabelOffset( 0.015,"Y");
RootStyle->SetLabelSize ( 0.050,"Y");
RootStyle->SetLabelFont ( 42 ,"Y");
RootStyle->SetTitleFont (42,"XYZ");
RootStyle->SetTitleColor (1);
// Options
RootStyle->SetOptFit (1);
RootStyle->SetMarkerStyle(20);
RootStyle->SetMarkerSize(0.8);
// cout << ">> Style initialized with the Root Style!" << endl;
// cout << ">> " << modified << endl << endl;
return RootStyle;
}
//need to get expected limit with sprinkling (counting exp and binned), observed limit given the two observed in the bins we saw them, observed limit including clustered events in passing events. try to do the same for dailey's?
|
| Attachment 2: plot_note.ps
|
|
11
|
Sun Mar 19 14:14:10 2017 |
Amy Connolly | Analysis | Analysis | Search for interstellar/interplanetary travel by alien civilizations | Other | See the attached papers. I wonder if we could look for these with ANITA and distinguish between natural and artificial origin.
Here are some estimates that I did last night.
The paper quotes that for an FRB at a distance ~10-20 kpc, we would see at S_nu=10^10-10^11 Jy where 1 Jy=10^-26 W/m^2/Hz.
For a BW=1 GHz and taking 1 m^2 effective area antennas, we get 10^-7-10^-6=V^2/R where R=50 Ohms, then taking the lower end of the range of S_nu gives V=2.2E-3 V.
For anita V(thermal noise) is roughly V_rms=sqrt(k_B*T*BW*R)=sqrt(1.38E-23 * ~340 K * 10^9 Hz * 50 Ohms) = 1.5E-5 V.
So let's say roughly we can hope to see signals down to the thermal noise level. Then as expected, an FRB in our own galaxy should be easily observable.
The paper estimates that in a galaxy there are ~10^-5 FRBs/day. Wikipedia tells me that within 3.59 Mpc there are 127 known galaxies. Given the observed voltage would go like 1/r, then the furthest ones from that group of 127 would be seen at voltages a factor of ~4 Mpc/20 kpc=200 lower, so the furthest ones would be right at the thermal noise level.
When ANITA is at altitude, we perform our searches below the horizontal, and can hope to see radio signals directly from horizontal down to the horizon at about -6 deg. So we view a sort of disk from 0 to 6 deg. in all directions in azimuth in payload coordinates. This intersects the galactic plane at some angle but we can not just look for FRBs in our own galaxy but also in the other galaxies which we pretend to be uniformly distributed in a spherical volume. If we could view from +90 deg. to -90 deg. then we would be able to see in all directions, so the fraction of the galaxy we can see is about 6/180=1/30. So the probability that over roughly 100 total days of livetime over all flights, ANITA sees an FRB can be estimated as:
Prob(seeing a FRB from within 3.59 Mpc)=10^-5 FRBs/day * 100 days * 127 galaxies * 1/30 = 0.004.
If the intensity received is on the upper end of the range given (S_nu=10^11 Jy), then maybe we can see out to 40 Mpc. Assuming the same density of galaxies as within 4 Mpc, since we're looking in a disk the # of galaxies goes like r^2, so we get a factor of 100 increase in prob-> prob=0.4, not bad.
But, we haven't accounted for the beam of the FRBs. It appears at first glance in their paper that power increases at lower frequencies, and the beam width increases at lower frequencies, I'm not sure, and those two things would both be good for us since FRBs have been observed above a GHz and so we could see them at lower frequencies.
Some things to look into:
Understanding their calculations of the beam width and power, and the frequency dependence
03/20: It seems the beamwidth is based on some multiple of the diffraction limit lambda/D where D is the size of the source.
How they are proposing one would be able to distinguish between natural and artificially produced FRBs
03/20: The beam would be shadowed by the sail, and maybe you would see fringes. Also a repeater could be a signature of the relatively short acceleration and deceleration phase.
Is ANITA unique is how much sky we can see at these frequencies with this sensitivity?
Would the fact that we digitize so quickly help in distinguishing natural from artificial?
Is there any time when ANITA is orientied in such a way that we look along the plane of the MW and thus see the whole galaxy?
Can we extend analyses above horizontal and increase sensitivity that way?
Can we look for reflections from the surface?
Note that the signals would extend ~1 ms in time. |
| Attachment 1: FRBs.pdf
|
| Attachment 2: Guillochon_2015_ApJL_811_L20.pdf
|
|
22
|
Wed Sep 13 09:28:15 2017 |
Amy Connolly | Analysis | Analysis | Info on generating pseudoexperiments, calculating likelihoods from them and finding p-values | Other | Will point to a bunch of papers and stuff here. |
|
13
|
Thu Mar 23 21:43:12 2017 |
Abdullah Alhag | Analysis | Analysis | The results from running Karoo on the inelasticity data | | See attatchd file. |
| Attachment 1: The_result_of_using_Karoo_on_inelasticity_data.pdf
|
|
20
|
Fri Jul 28 17:43:20 2017 |
Abdullah Alhag | Analysis | Analysis | GP algorithms | | In this post, I will be pointing out the advantage and the disadvantage of the GP algorithms I came across, particular Eureqa and HeuristicLab.
Eureqa is by far the fastest genetic algorithm software I came across. It is over simplified and easy to use. It has some built-in fitness function and also with some playing with the function that is being solved for and some other feature, it is possible for one to write his/her own fitness function. Moreover, the software is available for free for academic use and for most platform. Other features come with the software is the ability to normalize the data in different ways and even handle outliers and missing data. The program support a large collection of functions including trig and more complex one.
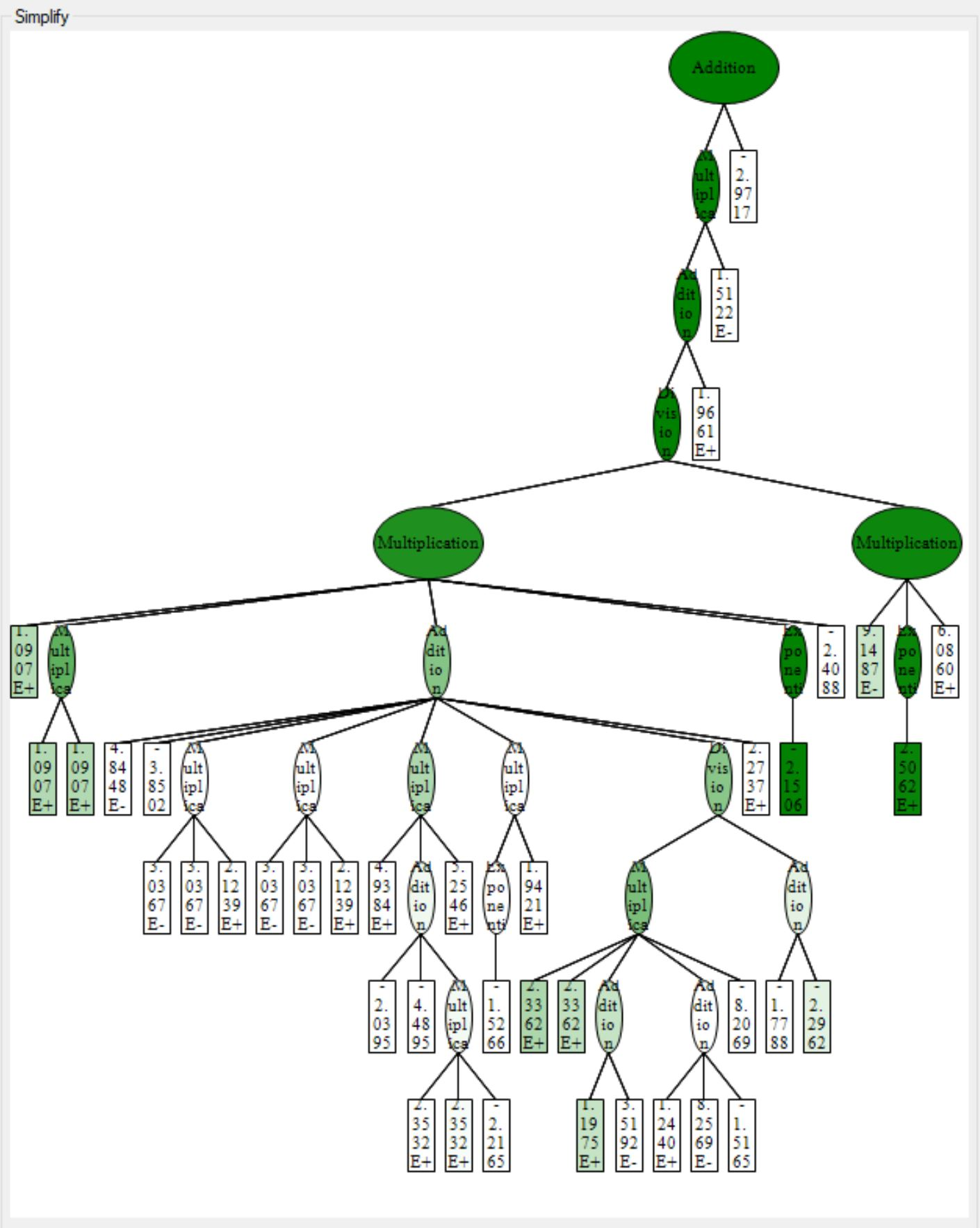
One the other hand, HeuristicLab is much slower than Eureqa but still far faster than Karoo-GP. The latest version of the software was released a year ago, and the support for the software is fairly slow. It is only supported for Windows; however, there is plans to adopted to Linux systems. The software support way more feature than Eureqa or karoo and even different regression and classification algorithms. You could also get the function ready to use in many software such as MATLAB, Excel, Mathematica, and much more. Another cool feature is that it shows you a three of the function and the weight of each node (operation or operand), greener means the node has more weight, see attached. It should be noticed that the software has the tendency to grow large three which could be fixed by changing the default max three length and the max three depth. The software has a problem with the last update of windows 10, you will get the blue screen if you opened too many windows, so be careful.
In booth software you could change how much of the data set goes to training and how much goes to testing, be sure to shuffle the data in HeuristicLab as it will otherwise distribute the data as training and testing non-randomly. Booth software by default shows plot of the current function with the x axis being the data entries (row numbers) and y axis being the target values with a curve of the function estimate values.
Below is a simple run of both software on a fake data that Prof. Amy gave me, see out.txt for data.
For start, using eureqa I got a few functions with an average of mean of R^2 of 0.98 or more which is very good.
First function: frequency = (571953.335547372*y + 15786079*x^2*y^4 + 297065746*x*y^2*asinh(x))/factorial(7.44918899410674 + x + y)
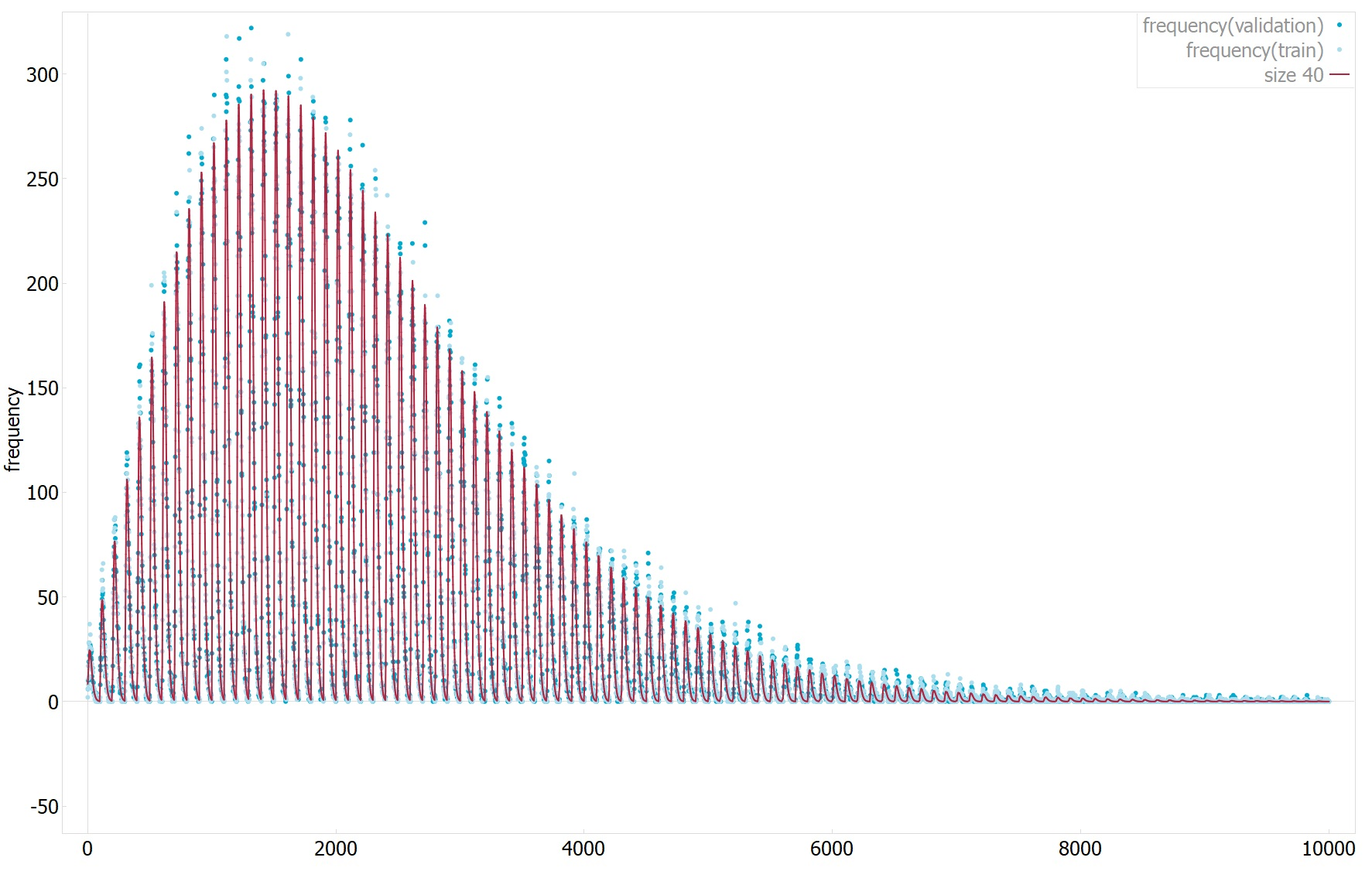
The first has an R^2 of 0.99(1 means perfect fit) and mean absolute error of 2.98(0 means perfect fit, data dependent, not normalized), see plot1 for 1D plot of the function estimate values vs target values.
Second function: frequency = (3569.91823898791*x*y - 149.144996501988 - 100.216589664235*x^2)/(5.26462242203202^x + 7.09216771444399^y*x^(1.3193170267439*x))
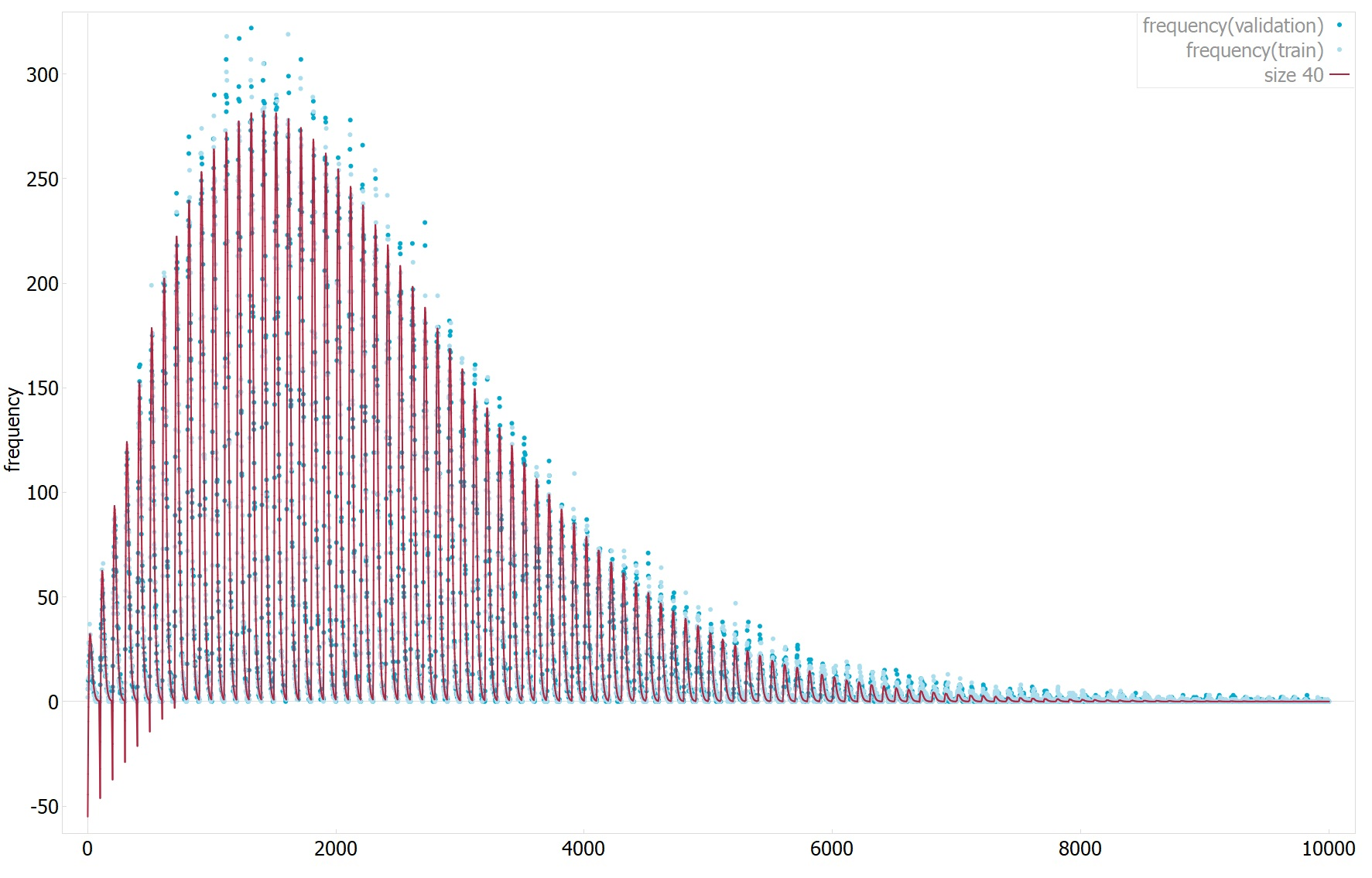
The second has an R^2 of 0.994(1 means perfect fit) and mean absolute error of 3.2(0 means perfect fit, data dependent, not normalized), see plot2 for 1D plot of the function estimate values vs target values.
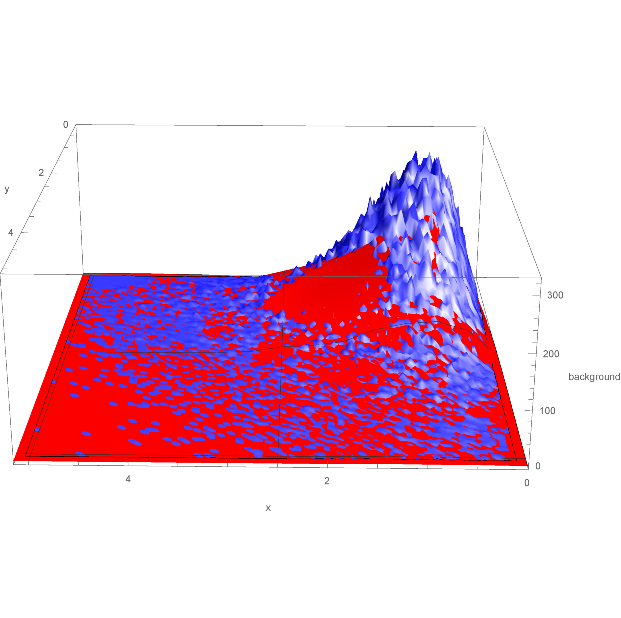
Also attached is 2D plot of the function against the data, the function plotted is the second function, but all are very similar, see Eq23.
Using HeuristicLab. The function below has an R^2 of 0.987, mean absolute error of 3.6 and normalized mean squared error of 0.012.
The function is: (((EXP((-1.3681014170483*'y')) * ((((-1.06504220396658) * (2.16142798579652*'x')) * (3.44831687407881*'y')) / ((((1.57186418519305*'y') + (2.15361749794796*'y')) / ((1.6912208581006*'y') / (EXP((1.80824695345446*'x')) * 16.3366330774664))) / ((((2.11818004168659*'x') * (1.10362178478116*'y')) - ((((-1.06504220396658) * (2.16142798579652*'x')) * (3.44831687407881*'y')) / ((((2.11818004168659*'x') + (2.15361749794796*'y')) / ((10.9740866421104 + (1.8106235953875*'y')) - (2.15361749794796*'y'))) / (((-7.8798958167) + (-6.76475761634751)) + ((2.87007061579651*'x') + (2.15361749794796*'y')))))) - (((((-8.85637334631747) * ((1.9238243855142*'y') - (1.01219957177297*'y'))) + (((-6.37085286789103) * 5.99856391145622) - ((-12.9565240969832) - 2.84841224458228))) - ((2.11818004168659*'x') * (1.10362178478116*'y'))) - (((2.11818004168659*'x') * ((((0.197306324191089*'y') + (0.255996267596584*'y')) - (2.16142798579652*'x')) - (((-1.06504220396658) * (2.16142798579652*'x')) / (12.2597910897177 / (1.25729305246107*'y'))))) - (EXP((-1.3681014170483*'y')) - ((((-6.29806655709512) * 6.39744830364858) / (12.2597910897177 / (0.728256926023423*'x'))) - (1.10362178478116*'y'))))))))) * 179.42788632856) + 2.24688690162535)
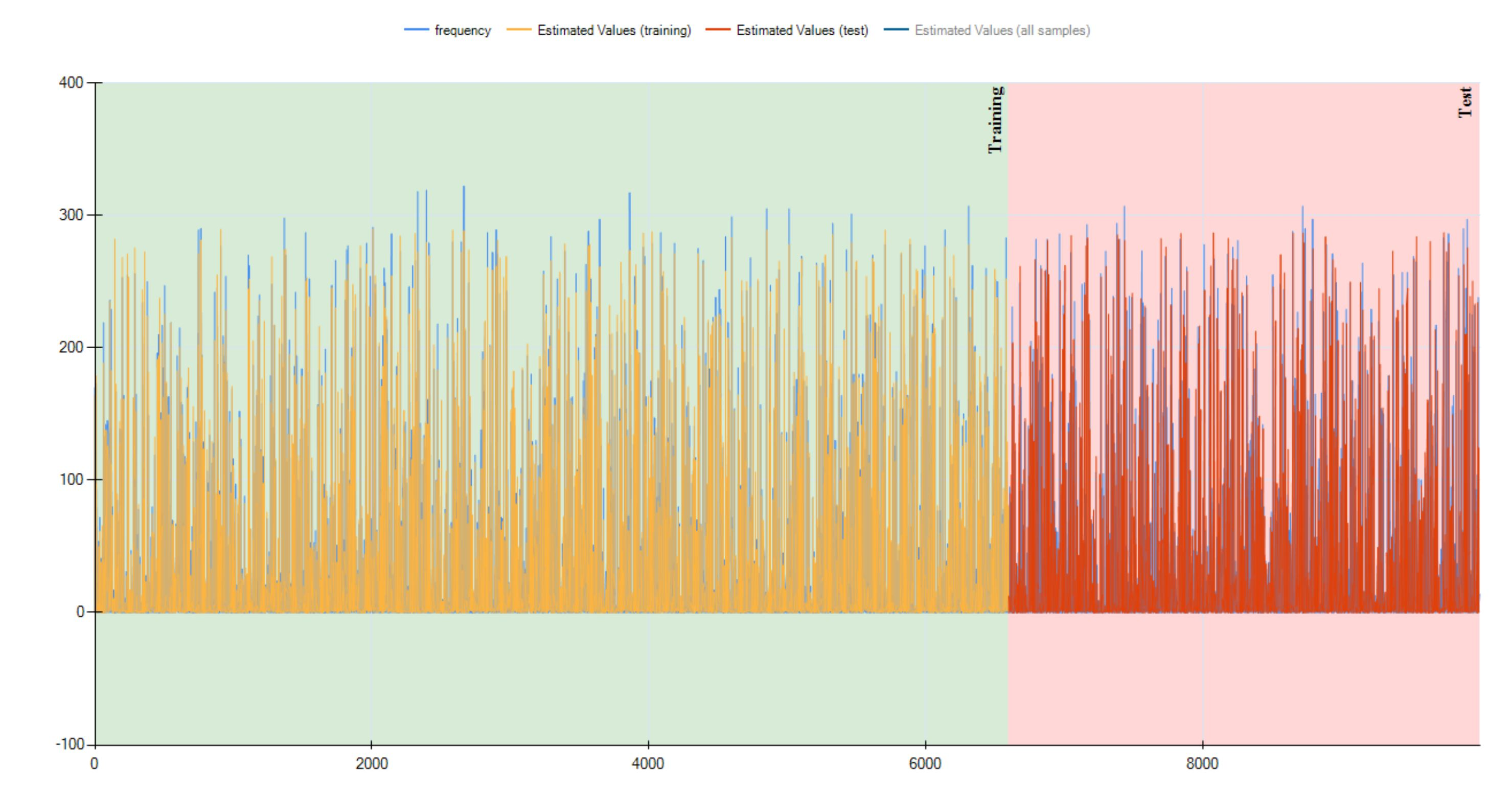
As you could see, HeuristicLab tend to generate function which are extremely large, this one has a depth of 15 and a length of 150.
See plot3 for 1D plot of the function estimate values vs target values, note that it is different from before because the data is shuffled, and three.jpg for the three representation of the function showing the wight of each node, greener means has more weight. |
| Attachment 1: plot1.png
|  |
| Attachment 2: plot2.png
|  |
| Attachment 3: plot3.png
|  |
| Attachment 4: Eq23-0.png
|  |
| Attachment 5: three.png
|  |
|
Draft
|
Fri Dec 16 11:24:09 2016 |
| Software Installation | | | | |
|