| |
ID |
Date |
Author |
Subject |
|
|
Draft
|
Tue Mar 24 15:38:50 2020 |
Julie Rolla | X11 Forwarding Issue for Windows | Most of our students
(1)Ubuntu doesn't come with X11 forwarding on its own. So you can install Xlaunch
xeyes -display :0
|
|
|
41
|
Wed Mar 18 15:01:10 2020 |
Julie Rolla | ONBOARDING: Tutorial Projects, Reading, Setup, etc. for New Students | For each new student joining the group, we ask them to onboard by completing each of the following items in the order given:
(1) Go to osc.edu and request an account
(2) Complete the following tutorials and reading. Note that the order of these does not matter, however, I would suggest "how to use Git" comes last, as it is the lease important for you to start. Finally, I advise you work on a tutorial concurrently with reading the pages assigned below from my Dissertation.
- OSC tutorials
- Because this project uses a lot of computation power, we run on the Ohio Super Computing Center's clusters. Please utilize these tutorials to learn more.
- Read Julie's Dissertation pages 1-9, 12-20 (starting at section 1.5.2), 28-37 (starting at section 2.3)
- Feel free to read chapter 3, as it has the history of this project and goes into details of our most up-to-date endeavors. This is optional and not mandatory.
- Complete the introduction to Bash project
- Complete the introduction to Python Project
- How to use Git (optional)
- Make sure to get an XFdtd license on OSC
(3) Finally, review the full manual and complete the bullets below. The full manual is Appendix A of Julie's Dissertation.
- Appendix A of my dissertation is our manual and acts as the holy grail of our research. Anything you need can be found here (from where our info exists on the web, to how to run the code).
- Once you have read this through entirely, we advise you log on to OSC and explore the code for PAEA in PAS1960. Once you have done so, complete this homework by making a copy of this google doc and filling it out on your own.
Once these are completed, the students are ready to start helping. This is all of the coding information and basic background information on evolutionary algorithms students must know to be proficient in assisting.
Other useful materials we have may include:
- Our old GENETIS manual
- This is somewhat out of date; however, it does have information on the general working on the loop.
- Julie's candidacy paper
- Complete the writing a genetic algorithm in C++ project
- This is still a work in progress! I'll complete it shortly!
- Read chapter 3 of this book (more if you have time) by Eiben and Smith, "Introduction to Evolutionary Computation" for a quick introduction. The book should be available electronically from a university library. Let us know if you can't find it there.
- Get to know XFdtd
- XFdtd manual is also attached; however, the link above (in 5) is more useful for coding a .xmacro.
Important notes:
MAC USERS: Using a GUI when connecting to OSC via SSH from your terminal.
You will need to set up X11 forwarding. Otherwise, when you go to plot via SSH, they will not display (they don't forward from OSC to your machine). If you are unable to run GUI's on OSC through your computer's terminal, follow these instructions:
- In your own system's terminal, type: sudo vi /etc/ssh/sshd_config
- Search for the following line: X11Forwarding no
- Change it to: X11 Forwarding yes. Save the file and exit.
- In your own system's terminal, type: sudo systemctl restart sshd
- For a Mac, download and install XQuartz.
- Connect to OSC using ssh -XY username@hostname.
- You can test that this works by typing 'xeye' or 'xclock' in the terminal when logged into OSC.
Now you should be able to use OSC's GUIs.
PC USERS:
As a PC user, there are a few things you need to do:
(1) Enable the Windows Subsystem for Linux on your device (https://developerinsider.co/stepwise-guide-to-enable-windows-10-subsystem-for-linux/#:~:text=To%20enable%20the%20Windows%20Subsystem,list%20here%20and%20click%20OK.)
(2) Get Bash on Ubuntu installed on your machine. You are going to need to have Bash on Ubuntu installed prior to starting these trainings; otherwise Bash commands will not be recognized.
(3) Get X11 forwarding working. Otherwise, when you go to plot via SSH, they will not display (they don't forward from OSC to your machine).
- Follow these steps here.
- Change your config file. To do so, in terminal (with Bash on Ubuntu open)
- type: sudo vi /etc/ssh/sshd_config
- Then change '#X11Forwarding no' to '#X11Forwarding yes' and save it.
- Make sure Putty and Xming are running every time before you ssh in. Use Xlaunch application to run Xming and press yes to everyrthing.
- Xming should now be running as a background task.
- Open PuTTY and open the pre-saved like you saved in PuTTY. (This will open a Bash terminal that asks for your OSC login password.)
|
| Attachment 1: 237799239-XFdtd-Reference-Manual-pdf.pdf
|
|
|
40
|
Tue Mar 17 16:18:29 2020 |
Julie Rolla | GENETIS Planning Meeting | Attendance: Julie Rolla, Alex M.
Today Alex and I met to discuss how to move forward given the COVID-19 restrictions. We have resent out a message polling people's schedules to create online working sessions. Here is what we are planning:
| Group name |
Tasks |
Participants |
Times per week to meet |
| Loop Group |
Running the loop, updating software, getting plots |
Julie, Alex M., Alex P. |
2 |
| Lecture |
Attend Leo's lectures |
Full group |
1 |
| Training (Python, c++, and Bash projects) |
Completing the training modules (Alex M. and Julie will be answering questions) |
Mitchell, Ryan, Evelyn, Eliot, Julie, Alex M. |
2 |
Once we collect times for each group, we will have separate meetings for all of these entities. This way it's easier for us to meet without all having different items to discuss/work on. All of these will be zoom meetings moving forward. Meeting info will be announced shortly!
Tasks for "the Loop Group":
- Make wall time a variable for XF sim.
- Test current software edits.
- Such as item #1 above
- Make sure the bottleneck function is working.
- Right now it seems to be working; however, we keep hitting a wall time error. If we can make the wall time a variable then we can be sure the bottleneck function is working, and the wall time is truly the error.
- Get Ruby working on XF
- when I type:
/usr/local/xfdtd/7.8.1.4/remcom/bin/xfdtd I get:
Info: Using latest available version (7.8.1.4)
Info: Running /usr/local/xfdtd/7.8.1.4/remcom/XFdtd_7.8.1.4/bin/xfdtd
Error: Unable to find executable to run. This command must exist
- Get Alex's account of Ruby working.
- He received an email and is still unable to log in. He is touching base with them again.
Tasks for "the Training Group":
- Julie and Alex need to finish the C++ project!
|
|
|
39
|
Fri Mar 6 15:01:37 2020 |
Julie Rolla | GENETIS Working Meeting | Attendance: Julie, Evelyn, Mitchell, Ryan, Alex M.
Since we have been having issues with the lack of availability of GPUs on OSC, we have come up with a new to-do list:
- Try to keep running on Pitzer with 1 core instead of 40. Based on first attempts, it is saying this will reduce wait times to ~3 hours (will vary based on usage), and after that 3 hours, human interaction will be needed. So the plan is, someone requests a job in the morning, and when the interactive job starts, the person is responsible for doing the thing which will allow the generation to run. We will aim for one generation per day under this plan, taking turns. Better than no evolving.
- I have asked Heechang at OSC if we can install the latest version of XF on Ruby and/or Owens. That would be better. There seems to be more available on Owens at least.
- Our initial attempt to run on Ruby is giving us an error which is probably to do with different inputs required in different versions. Will attempt to pursue this as a last resort, but the older version of XF does require lots more human interaction so until other options are exhausted it's not worth it.
Today's Tasks
Mitchell: Worked on Python Project, learned to evolve (note: Mitchell may leave the group at the end of the semester)
Ryan: Starting the Python project
Evelyn: Finish bash project.
Julie: C++ project, look at Amy's proposal draft.
Alex M: Fix XF solver to submit each individual as a separate job (thus we can request a smaller wall time for each job)
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF.
- We are going to pause on working on this, because we are struggling to get GPU time. We will test this whenever we see OSC less busy (ie if we log on at a lucky time).
- Evolve: Try to keep running on Pitzer with 1 core instead of 40. Based on first attempts, it is saying this will reduce wait times to ~3 hours (will vary based on usage), and after that 3 hours, human interaction will be needed. So the plan is, someone requests a job in the morning, and when the interactive job starts, the person is responsible for doing the thing which will allow the generation to run. We will aim for one generation per day under this plan, taking turns. Better than no evolving.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
As of 3/5, I have not gotten an update on this. I will check on the status of this.
-
Comment code thoroughly (Everyone needs to help with this)
-
Julie made some progress on this over the weekend.
-
Made comments on what each bash script version does, and cleaned up our directories from old code so that it doesn't confuse the others when they start helping
-
WAY more needs to still be done.
-
Get new proposal results for March 12th deadline:
-
Julie is looking at what Amy wrote here
|
|
|
38
|
Wed Mar 4 01:00:35 2020 |
Julie Rolla | GENETIS Working Meeting 03/03/2020 | Attendance: Julie, Mitchell, Ryan, Alex P., Leo, Alex M.
Today's Tasks
Mitchell: Worked on Python Project and helped with Ryan's final bash project questions
Ryan: Finished the bash project, and is about to start the Python project.
Leo: Worked on lectures, which will restart on March 17th.
Julie, Alex P, Alex M: Worked on fixing the job XF submission scripts. Worked on making it so that it saves state after each individual's XF simulation is complete. We tried to parallelize as many of the XF scripts as possible, but if we have more than one it asks us to choose a license for each individual -- ie requires user input each time. Since we haven't found a way to get around it AND since running with 1GPU is super fast (under 5 min per individual), we are going to run them in series.
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF. (PASSED THIS TASK TO JULIE. WILL DO TOMORROW 3/4)
- Evolve: Right now, we are using an interactive job with only a CPU running at half scale, and have just edited our software so that we submit jobs for the XF simulations requesting 1GPU (could be 2GPU if we choose to edit it so).
- WHY WE ARE DOING THIS:
- Remember that we only need an interactive job when XF initializes and reads in the DNA for each individual. The GUI closes once the simulations start and we no longer need an interactive job.
- We were requesting a 1GPU interactive job and just running XF from the command line; however, OSC has been SUPER busy lately and we haven't been getting them (especially since we need long wall times if we are running the full simulations over many generations within the interactive job). However, we realized that if we submit an interactive job at a CPU (so that XF can allow the GUI to pop up), and then run the simulations by requesting 1-2GPUs we can then request a much smaller wall time and it will be quicker for the simulations to run --- ie we would be requesting a wall time of about 10 minutes using 1-2GPUs per simulation for each job submission versus 5+ hour wall time of an interactive job using 1-2GPUs.
- WE HAVE TESTED THIS, AND WE HAVE ERRORS. WE WORKED ON FIXING THEM TODAY, AND HOPE TO FINALIZE THEM TOMORROW (3/4) AND RUN!
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
As of 3/3, I have not gotten an update on this. I will check on the status of this tomorrow.
-
Comment code thoroughly (Everyone needs to help with this)
-
Julie made some progress on this over the weekend.
-
Made comments on what each bash script version does, and cleaned up our directories from old code so that it doesn't confuse the others when they start helping
-
WAY more needs to still be done.
-
Get new proposal results for March 12th deadline.
|
|
|
37
|
Sat Feb 29 02:21:16 2020 |
Julie Rolla | GENETIS Working Meeting | Attendance: Julie, Mitchell, Ryan, Evelyn, Eliot, Alex M.
Today's Tasks
Julie: Worked on C++ project, and helping others with the Python project; I also made additions, clarifications, and edits to the Bash project (also updated the solutions and pushed them to gitHub). I still need to do the following:
- Add the instructions from the google doc to the GitHub pages for both the Python and the Bash projects.
- Work on the C++ project!
- Run a test evolution this weekend to make sure AraSim stuff is fixed and that XF job submissions work.
- EVOLVE
Mitchell: Started Python Project.
Evelyn, Eliot, Ryan: Work through my bash project.
Leo: Worked on lectures.
Alex M: Helped make XF job scripts. Needs to do the following:
- Parallelize the AraActualBicone properly.
- EVOLVE!
Alex P: Needs to do the following:
- Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF.
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF. (ALEX P)
- Evolve: Right now, we are using an interactive job with only a CPU running at half scale, and have just edited our software so that we submit jobs for the XF simulations requesting 1GPU (could be 2GPU if we choose to edit it so).
- WHY WE ARE DOING THIS:
- Remember that we only need an interactive job when XF initializes and reads in the DNA for each individual. The GUI closes once the simulations start and we no longer need an interactive job.
- We were requesting a 1GPU interactive job and just running XF from the command line; however, OSC has been SUPER busy lately and we haven't been getting them (especially since we need long wall times if we are running the full simulations over many generations within the interactive job). However, we realized that if we submit an interactive job at a CPU (so that XF can allow the GUI to pop up), and then run the simulations by requesting 1-2GPUs we can then request a much smaller wall time and it will be quicker for the simulations to run --- ie we would be requesting a wall time of about 10 minutes using 1-2GPUs per simulation for each job submission versus 5+ hour wall time of an interactive job using 1-2GPUs.
- WE HAVE NOT TESTED OUR .SH JOB SUBMISSION SCRIPTS! JULIE WILL DO IT THIS WEEKEND!
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
Comment code thoroughly (Everyone needs to help with this)
-
Get new proposal results for March 12th deadline.
|
|
|
Draft
|
Tue Feb 25 17:10:36 2020 |
Alex Machtay & Alex Patton | Working Meeting 2/25/20 |
Today's Tasks
Mitchell, Evelyn, Eliot, Ryan: Work through the bash project; Mitchell is done, Ryan and Evelyn are close to being finished.
Leo: Worked on next presentation (not sure when he'll present--either Friday or next Tuesday).
Alex M: Continued commenting code and helped Alex P. with changes in Part_B and Roulette_algorithm .
Alex P: Fixed errors in loop, tested with an interactive job (using CPU). Errors look to be fixed up through part B. Arasim should be launching and using the $tmpdir, though we need to get further in the testing to see for sure.
Made a copy of the loop that functions without GPUs so that if GPU is unavailable some progress can be made even if it is slow.
While waiting for a GPU Alex P ran on CPUs, at half scale factor one indiviudal in XF took 32 minutes (although it was predicted to be over 2 hours).
|
To Do List:
We want to keep testing the loop. Right now, we are using a cpu running at half scale. This gives estimated times for each antenna of up to 2 hours. GPUs are suddenly hard to come by, so if we are relegated to using CPUs we'll probably need to scale down considerably.
Evelyn, Ryan, and Mitchell are probably going to be ready for the python project soon. If someone is around on Friday to help them they should be able to finish the bash project.
One idea is to run all the inital xmacros in the interactive job and then submit a GPU job that runs all the xfsolver command. With the hope that not having the GPU in the interactive job will make it easier to get the interactive job and to get the GPU for a shorter time because it would only be needed for a smaller segment. This would especially help with running larger amounts of individuals.
|
|
|
35
|
Fri Feb 21 16:47:21 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: I finished the Python assignment. I am making sure my solutions encompass all of the different ways to solve this, and will then start on the C++ project.
Mitchell, Evelyn, Eliot, Ryan: Work through my bash project.
Leo: At 4 pm today Leo will begin his "physics of antennas" lecture series!
Alex M: All scaling is corrected to be set at the bash script level. Alex is now going to work on commenting code!
Alex P: Still working on our AraSim edits!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
UPDATE: Alex still trying to figure out how to input Car's suggestion. He also needs to add this for the flag files that are being saved. Here is his most recent question:
Alright so I have pbsdcp working with generic files and I now have it working in the AraSim call but my current worry is that it wasn't creating a file unless I had
./AraSim setup.txt $runNum outputs/ a_${num}.txt > $AraSimDir/outputs/a_${num}.txt
So I just did this to have some a_1.txt, before we had the last line writing directly to our antennaPerformance metric (this makes the AraOut file typically I just kept it as this name for testing purporses), now if I get rid of the portion after the > or the part after outputs/ it doesn't seem to make a file that we need. I think I don't understand to get the full output of AraSim without using this. Because I don't understand how using this is and then using pbsdcp is any more efficient.
Now maybe if I had the portion right after the > go directly into TMPDIR instead of using pbsdcp to both scatter to and gather from TMPDIR that would be better
-
Make the following variables in the bash script.
-
UPDATE: Alex M. finished editing the scaling factor to be at the bash level for every piece of code that uses it.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
UPDATE: Alex commented code that utilized the scale factor (Part_E.sh). He is now going to begin commenting the other code to clean it up. We want it to be read as cleanly as our GA.
|
|
|
34
|
Tue Feb 18 16:20:59 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Continue writing Python project!
Mitchell, Evelyn, Eliot (not here on Tuesdays), Ryan: Work through bash project
Leo: continuing gathering information for the "physics of antennas" lecture series! We start this Friday at 4pm.
Alex M: Try to get scaling into the xmacro, comment the code better.
Alex P: Add on the AraSim fixes mentioned below! He is testing this now. We are waiting for updates.
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
UPDATE: Alex P. has edited our bash job script to save the outputs to $TMPDIR and then to pbscp (submit a "job" to copy them over) them into our correct directory that the bash script needs them in. He is testing this now and will report back.
-
Note: we still need to make sure that every time we run that we divide AraSim runs into MANY more jobs (ie further parallelize by dividing into, say, 100 instead of 10).
DATA_LIKE_OUTPUT=1,2 doesn't work with DETECTOR=0,1,2
DATA_LIKE_OUTPUT controls data-like output into UsefulAtriStationEvent format; without a real station selected (using DETECTOR==3,4), the mapping to the data-like output will not function correctly
There are 1 errors from settings. Check error messages.
SysError in <TFile::Flush>: error flushing file outputs/AraOut.setup.txt.run29.root (Stale file handle)
- Explanation of issue (2): We keep timing out of the interactive job when AraSim is running (if the user isn't remaining active on their terminal).
- OSC suggested that we use a virtual desktop (via ondemand) instead of using an interactive job. We will try this.
- Note: We should now be requesting 2GPUs instead of 1GPU. UPDATE: Alex P. requested one, but it took too long and he had to leave before he got it.
-
Make the following variables in the bash script.
-
UPDATE: Alex M. figured out how to pass the scale factor into the xmacros. We used the "tr" command in bash. He will be testing this and can update us at the Thursday meeting.
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
UPDATE: Alex commented code that utilized the scale factor (Part_E.sh). More still needs to be done.
|
|
|
33
|
Fri Feb 14 16:44:44 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Write Python project! I have officially finished the bash course for the students. It can be found here: https://docs.google.com/document/d/1nGdPrwfYJOrO6NBT76Wsh6T0hDmGHo2-23oJ1WyeNcs/edit?usp=sharing. Mitchell is currently trying it out and will give me comments.
Mitchell: Work through my bash project and give me comments!
Evelyn, Eliot, Ryan: Make progress on the "Learn the Command Line" course on Code Academy.
Leo: continuing gathering information for the "physics of antennas" lecture series! These will take place every Friday meeting starting next week.
Alex M: Try to get scaling into the xmacro, comment the code better.
Alex P: Add on the AraSim fixes mentioned below!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
Carl believes this is due to the way we are saving our AraSim outputs. Instead of saving the files directly to our desired location, we are going to save them to $TMPDIR and use "pbsdcp" to copy them from $TMPDIR to our desired location. This may alleviate the problem since it will not be writing all the files to our desired location at once.
DATA_LIKE_OUTPUT=1,2 doesn't work with DETECTOR=0,1,2
DATA_LIKE_OUTPUT controls data-like output into UsefulAtriStationEvent format; without a real station selected (using DETECTOR==3,4), the mapping to the data-like output will not function correctly
There are 1 errors from settings. Check error messages.
SysError in <TFile::Flush>: error flushing file outputs/AraOut.setup.txt.run29.root (Stale file handle)
- Explanation of issue (2): We keep timing out of the interactive job when AraSim is running (if the user isn't remaining active on their terminal).
- OSC suggested that we use a virtual desktop (via ondemand) insead of using an interactive job. We will try this.
- Note: We should now be requesting 2GPUs instead of 1GPU.
-
Make the following variables in the bash script.
-
Alex M is working on this. We can't directly make this a variable. We need to take the array of frequencies and multiply it by some value and THEN feed it into the xmacro. He is looking into whether of not he can directly do this in bash. If not, I have written a small python script that does this, and he can write it to that file; however, he can't append it at the end so it's not the ideal way. We are going to continue to try to do it through bash first.
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
The Gen alg is commented so thorougly that it can read like a book. We want this with ALL of our code; it will help all the less familiar people get comfortable.
|
|
|
32
|
Wed Feb 12 01:18:22 2020 |
Julie Rolla | GENETIS Working Meeting | Today we continued with the to-do lists items previously posted. I marked all the to-do list items that have been completed with green and a slash. Note that I have also added a new item at the bottom of the to-do list.
Today's Tasks
Julie: Write bash practice assignment (almost done!)
Evelyn, Mitchell, Eliot, Ryan: Make progress on the "Learn the Command Line" course on Code Academy. (Mitchell is done, and the others are close.)
Leo: Start gathering information for the "physics of antennas" lecture series! Note: Remember that Leo wants to be an educator, and would like us to find opportunities to work on "teaching" items. Most of the students don't have any background on how antennas work. He started learning about them and is going to do a lecture series
Alex M: Make sure antenna scaling is made a variable in the bash script everywhere that it exists, start mass commenting all of the code so it reads as cleanly as the GA does. (Finished making the antenna scaling a variable. needs to be tested, but we waited forever for wall time)
Alex P: Fix hard-coded variables (done but tried to test it. Waited for wall time and never got it), and looked into ways to get around requesting an interactive job. For some reason, we don't need an interactive job to forward things like plots, but we do for the XF UI. We were trying to figure out why this is, and if there's a way to fix this. So far, no luck.
AMY QUESION: Could we actually consider running on NASA computers? I think Alex P. found a way the NASA computing centers let you adjust for this issue, and it didn't work on OSC. If we end up not finding a solution with OSC, is it at all possible to have XF installed with NASA computing centers? (NOTE: I JUST ASKED ALEX FOR MORE DETAILS AND WILL ADD THEM HERE TOMORROW -- IE WEDNESDAY)
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
-
Comment code thoroughly
-
The Gen alg is commented so thorougly that it can read like a book. We want this with ALL of our code; it will help all the less familiar people get comfortable.
|
|
|
31
|
Mon Feb 10 14:20:56 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Write bash practice assignment, assist others when they have questions, work on Git lecture. (Will finish bash assignments early next week)
Evelyn, Mitchell, Eliot: Make progress on the "Learn the Command Line" course on Code Academy. (Should be done by the end of the week?)
Leo: Start gathering information for the "physics of antennas" lecture series! Note: Remember that Leo wants to be an educator, and would like us to find opportunities to work on "teaching" items. Most of the students don't have any background on how antennas work. He started learning about them and is going to do a lecture series (likely the first 20 minutes of every meeting) starting in a few weeks! (Should start series by hopefully 2/21)
Alex M: Clean up unused code. Fix hard-coded directories (likely finished & tested by end of day 2/11)
Alex P: Fix hard-coded variables (likely finished & tested by end of day 2/11)
I've tacked on our to-do list in the meantime so we can keep carrying the items still pending along with us. As we complete them, I will drop them off the list!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables.
-
Xmacros (use sed command)
-
All over bash script functions.
-
AraSim job files (sed command?)
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
|
|
|
30
|
Tue Feb 4 17:17:10 2020 |
Julie Rolla | GENETIS working meeting | As of 01/30/2020, the proposal was submitted! The project description and summary are attached below.
Bicone Loop Status Update:
The loop currently runs; however, it does seem that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
The loop currently has the following additions implemented.
- Functionalized loop (XF_Loop_AraSeed.sh)
- Cleaner bash script which runs smaller bash scripts, instead of one massive script.
- Submits multiple AraSim jobs in parallel for each individual to increase speed.
- Ie if 100k neutrinos are thrown per individual ($NNT=100000, $Seed=10) we can, say, break it up into a "seed" of 10 -- which then submits 10 jobs with NNT=10k for each individual.
- Still needs this to be fixed: The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded. It is just submitting one job the following way.
- If NNT=100000 and Seed=10 (ie 10 jobs with 10k neutrinos), it will only run AraActualBicone as 1 run with 10k neutrino number (ie NNT/seed=NNT_New).
- Save State
- After every "part" -- ie part a, part b, etc -- of the loop, we save state by writing a series of numbers to a text file.
-
If the loop is killed for any reason during the process, the text file is read in, and the loop can continue forward from the last completed item/section.
-
This saves the state after every individual runs through XF.
-
Antenna sizing
-
The larger the antenna, the longer XF takes to run. A true-to-size ARA antenna takes anywhere from 1.5 to 2 hours per individual.
-
What we've done: Allowed us to scale the antenna by a factor of "x".
-
If we make the gaussian centered around something, say, 5 times smaller than the ARA antenna size, we then make the frequencies 5 times larger. This should have the same effect.
-
This also means we've had to scale these sizes back up when plotting.
-
Still needs this to be fixed: all of this has been hardcoded in! We need to make these variables in the MANY places we need to adjust for this scaling. See the to-do list below for details.
Moving forward, I will be doing the following:
- Getting our less coding-savvy teammates familiar with bash, python, and c++. This will be crucial for us when we start adding more parameters to our GA.
- Mastering XF (along with Alex, and Alex).
- Getting the team to clean up some messy coding done during the proposal hustle.
- Fixing some errors.
- Teach the ENTIRE team git.
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables.
-
Xmacros (use sed command)
-
All over bash script functions.
-
AraSim job files (sed command?)
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
|
| Attachment 1: project_description.pdf
|
| Attachment 2: project_summary.pdf
|
|
|
29
|
Thu Jan 9 14:24:25 2020 |
Julie Rolla | Winter Break Updates and new to-do list (with deadlines) | The following items from our to-do list have been successfully completed. Below this list, you can find the remaining urgent items for our proposal due date of 1/28 (items must be completed with plenty of time to spare).
Completed items:
*Note* please check the highlighted item. This one still must be verified.
- Made software plot Theta VS Gen
- Fixed current L and R plots
- These now have correct axis labels, a legend, and connect data for each individual between generations.
- Produced Veff for ARA's actual bicone parameters.
- Made XF_Loop.sh submit a job for the ARA gain AraSim input file in BiconeEvolutionOSC/AraSim/Ara_Bicone6in_outputs.txt
- STILL MUST: Make sure that the "wait function" has been properly edited to consider the submission of this extra job in the first iteration of the loop. (Not sure if $NPOP+1 instead of $NPOP fixed it correctly) ----> CADE SHOULD LOOK AT THIS :)
- Added plotting software for Veff VS Generation (includes ARA's actual bicone Veff as generated in #3).
- Fixed the ARA hole constraint in fitnessFunction_ARA.cpp -- ie fixed the calculation to consider the correct radius "r". See http://radiorm.physics.ohio-state.edu/elog/GENETIS/28 for details.
- Successfully made energy a variable in setup.txt.
- used the same "sed" format as before in the bash script (XF_Loop.sh).
- Pushed to Github.
Items to still do:
- Save CAD models in XF: Due 1/15 ------>(Cade-- second most urgent one)
Stop plots from popping up during run: Due 1/15 ------>(Alex P.)- Add "save state" idea to loop: Due 1/15 ------> (Cade and maybe Alex P can help -- This is the most urgent one)
- Figure out why fitnessFunction_ARA.cpp isn't plotting: Due 1/15 ------> (Julie, and Alex M)
- Run the loop and evolve for results: Due 1/24 ------>(Alex M, Mitchell, Cade)
- Write proposal: Final Due 1/28 ------> (Amy, Kai, Julie)
When submitting plots, please make a new folder in there and label it "Date_of_run, Energy, Generations, Individuals". For example, my folder within this dropbox link would be: "1_10, 10^19, 10, 5". I have made sample folders in this dropbox link if needed.
You can export the plots from OSC to your machine (using CP command) or can type "display NameOfPlot.png" then screenshot and crop your plot to upload to dropbox. |
|
|
28
|
Thu Dec 5 16:58:55 2019 |
Julie Rolla | Updated To-Do List: | Updated To-Do List:
- Add ARA constants in plotting code: ie add Veff, theta, R for ARA. This will sit as a constant line for us to compare each evolving parameter to. For example, in the plot Veff vs Generation, we will see a constant line for ARA's Veff to see how it is evolving in time.
TASK 1: MAKE Veff PLOT. Note: One of our variables in this run is Energy. Unfortunately, we care about how Veff changes with Energy for ARAactualBicone. This means that before plotting "Veff versus Generation" we need to generate Veff for ARAactualBicone at the energy we are assigning the run.
STEP 1: Make plotting code to plot "Veff vs Generation".
- You can follow the way we make LRPlot.py.
- However, in this case, we will need to take the files /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Ouputs/$RunName/AraOut${gen}_${i}.txt -- ie the AraSim outputs for each individual in ALL generations. ie Gen 0 - ${gen} -- INCLUDING the one we created in Gen 1 for ARAactualBicone -- and plot Veff from those.
- We also want to plot error bars with Veff. The error bars for each are labeled "and Veff (water eq)" at the bottom of these AraSim output files we are already reading in for Veff.
- Note, to know how to read in Veff, you can look at how we are reading that *exact* line from the AraSim output files in on the fitnessFunction_ARA.cpp code in the Antenna_Performance_Metric directory, since we are doing it there.
- Note that since we are plotting "Veff vs Generation", we will either have to read in all the files AraOut${gen}_${i}.txt (up to how may ${gen} we are in) every time -- including the ARAactualBicone one-- or we can append them all to one .txt file. It's your choice.
- Make sure you label the units for each axis every time, and that you have a key! If not, Amy will hate the plot!
STEP2: Add Veff for ARAactualBicone as a constat in the newly made "Veff vs Gen" plotting code. Remember, as I mentioned above, we must first put the ARAactualBicone through AraSim to see Veff at the same energy as the rest of the individuals. Here's what to do:
- Edit the Bash script to submit a job that runs the gain patterns of ARAactualBicone through AraSim in the initial gen (gen 0)!
- There's a current file that exits for ARAactualBicone that has the gain patterns of the current antenna, in the correct format for AraSim to run. In the bash script (XF_Loop.sh) we need to create ONE more AraSim job submission for this antenna (as a comparison to what we see with our genetically evolved ones) just in gen 0 (the first generation).
- This current AraSim input file for ARAactualBicone exists in BiconeEvolutionOSC/AraSim/Ara_Bicone6in_output.txt
- This needs to be the input for our .job script.
- Add lines to your newly made "Veff vs Gen" plotting code that reads in the Veff output for this ARAactualBicone AraSim output file (whatever you've chosen to name it when you wrote it into the job script.
STEP 3: Move the ARAactualBicone output file into the $Run_Name directory so it is not overwritten ever.
- We will want to make sure this is not overwritten after the run ends, and we start a new one.
- We will also be using it for plotting "Veff vs Generation".
STEP 4: Correct the "wait" command for the AraSim job submissions.
- The "wait" command, on the first iteration, will have to wait for $NPOP+1 now-- not $NPOP.
TASK 2: MAKE "Theta vs Gen" Plot.
STEP 1: Make plotting code to plot "Theta vs Generation".
- You can follow the way we make LRPlot.py; however, MAKE SURE YOU LABEL AXIS WITH UNITS, AND HAVE A KEY.
- This will be exactly the same as this other code, except you will need to:
- Read in theta from your generation DNA files
- Add the ARAactualBicone constant to the code as a reference point
TASK 3: Fix "L vs Gen" and "R vs Gen" plot.
STEP 1: Correct both plots to have labeled the units for each axis every time, and a key. This is plotted in the "LRPlot.py" code.
STEP 2: Fix our 'R' values. I do believe we are plotting the incorrect "R". Here is what I think is happening:

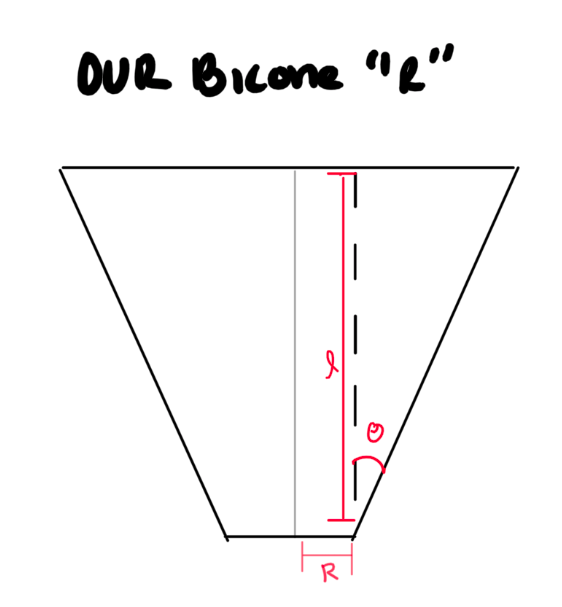
- We need to double-check and see what ARA defines "R" as on their current antennas.
- Check with Amy?
- What does Brian Clark's thesis say?
- Can you find a paper that shows a picture of how they define it?
- If we are wrong, this means we need to change the "R vs Gen" part in the "LRPlot.py" code so that we:
- Read in R, L, and Theta for each generation.
- See how this is already done for reading in R, and repeat it for L and theta.
- Do trig to calculate the same "R" as ARAactualBicone defines. See picture below!
- Store that newly calculated "R" as an array, and plot THAT.
- Don't forget to add the ARAactualBicone Radius as a constant line.
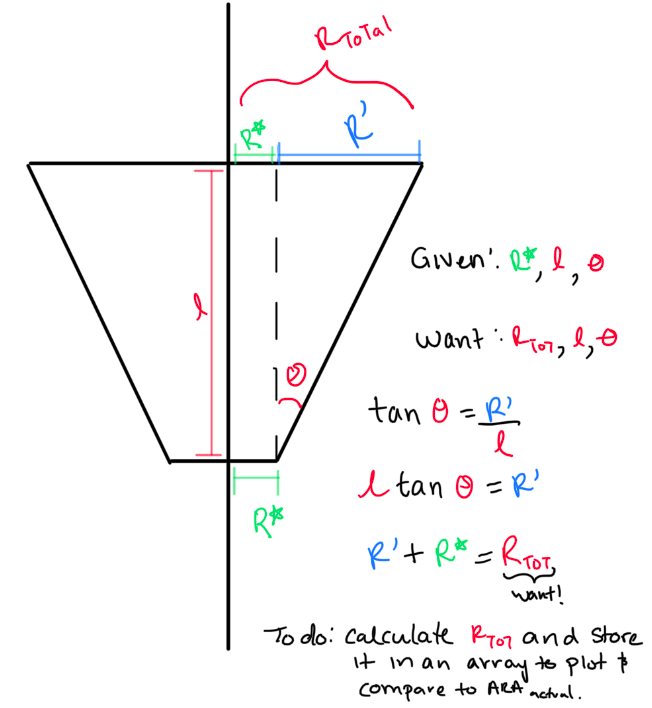
TASK 4: See PDF for other items! |
| Attachment 3: Proposal_To-Do_List.pdf
|
|
|
27
|
Thu Dec 5 14:32:54 2019 |
Julie Rolla | Adding Constraints To-Do List | In my last post, I mentioned (1) our to-do list for adding the ARA hole constraints, and (2) that we've made a mistake upon implementing it. I've attached detailed steps of that to-do list that I've written, as well as an explanation for the correction that is currently being worked on. Please see the pdf attached for an explanation of corrections.
|
| Attachment 1: AddingConstraints.pdf
|
|
|
26
|
Tue Dec 3 15:26:42 2019 |
Julie Rolla | Working session 12/3/19 | Attendance: Julie, Alex M., Alex P., Cade, Scott.
What we have done today:
(1) Fixed Concerns regarding copying the output .uan files properly so they are not overwritten. (see #8 in this elog entry: http://radiorm.physics.ohio-state.edu/elog/GENETIS/22 for details on the error). This was corrected in part E as:
for i in `seq 1 $NPOP`
do
for freq in `seq 1 60`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${i}_${freq}.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}_${freq).uan
done
done
And in the "looping part" -- ie part G we have corrected it to say the following. Note that every time we run an iteration/generation in the loop, it writes the name of the output files the exact same (individual_frequencystep.uan). It will overwrite these files in the next generation unless we move them and rename them like so. We don't want this, because we will likely want to plot the gain patterns of the individuals we like (gain pattern data is in the .uan files).
for i in `seq 1 $NPOP`
do
for freq in `seq 1 60`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${i}_${freq}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}_${freq}.uan
done
done
(2) Added scale factor "A" (actually called "Scale Factor" in our bash script and fitnessFunction_ARA.cpp), and the generationDNA.csv as an input file (this file is where the radii of all individuals in the current generation live).
- We have added this as a variable in the fitnessFunction_ARA.cpp. It now runs as a flag pass in when it runs in the bash script. In section E:
./fitnessFunction.exe $NPOP $ScaleFactor $AntennaRadii/generationDNA.csv $InputFiles #Here's where we add the flags for the generation
and in the looping part (part G):
./fitnessFunction.exe $NPOP $ScaleFactor $AntennaRadii/generationDNA.csv $InputFiles #Here's where we add the flags for the generation
and at the top where variables are declared:
RunName='Julie_12_2_2' ## Replace when needed
TotalGens=6 ## number of generations (after initial) to run through
NPOP=4 ## number of individuals per generation; please keep this value below 99
FREQ=60 ## frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=10000 ##Number of Neutrinos Thrown in AraSim
ScaleFactor=1.0 ##ScaleFactor used when punishing fitness scores of antennae larger than holes used in fitnessFunctoin_ARA.cpp
Note that every time the roulette algorithm runs, it's written into generationDNA.csv. We copy those right before rerunning the roulette alg in the next generation -- ie cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv -- so that each generations parameters don't get overwritten. However, the current generation's parameters live in generationDNA.csv until the roulette alg runs in the next generation, so we can read in that file to the fitnessFunction_ARA.cpp file to determine the constraint (ie Rantenna)
(4) Implemented Alex P.'s version of his optimized AraSim. This is now fully functional in the loop.
NOTE: WE HAVE MADE A MISTAKE! WE MADE THE CONSTRAINT THE R-VALUE WE READ IN FROM THE GENERATIONDNA.CSV FILE. THIS IS NOT THE RADIUS WE THINK IT IS! WE NEED TO DO SOME TRIG ON IT TO FIX IT!!
|
|
|
25
|
Tue Dec 3 01:26:52 2019 |
Julie Rolla | Working session 12/2/19 | Attendance: Julie, Alex P., Cade, Alex M., Mitchell, Evelyn (Go team!)
I. Adding Constraints on the in-ice hole size.
Today we worked on adding constraints on the drilled hole size. Our plan is to do so by penalizing the fitness scores of indiviuals with a radius larger than that of the hole in the ice. Ie in fitnessFunction_ara.cpp we would have:
if Rantenna>=R hole
fitnessScore =fitnessScore x (e-[A(Rantenna-Rhole)/Rhole)]^2)
else
fitnessScore= veff (or whatever way we decide to calculate the fitness score in the future, if this ever changed)
Where "A" is a constant we can adjust to tweak how heavy the penalty is. Other things to do in order to make this work:
(1) Make constant "A" a variable in the bash script.
- Do this the same way $NPOP is used in the roulette algorithm cpp program! :)
(2)Read in the roulette algorithm output file for your radius of the antenna.
- Note that this isn't a super obvious thing. Right now our roulette alg outputs are saved as $(generation)_filename.csv. So, for the first gen -- ie gen 0-- it will be 0_filename.csv. For the second gen -- ie gen 1-- it will be 1_filename.csv...etc. This means we need to make sure the fitness score calculator has a way of knowing what generation we are on, in order to be sure it is grabbing the correct radius for each individual (not that of an old generation).
We will be working on this more in our meeting tomorrow. As for now, the game plan has been to make comments on what we should be adding in the correct sections of fitnessFunction_ara.cpp (and will be doing the same in the bash script shortly, too --ie XF_Loop.sh).
II. Adding Ara Values to Plot.
We haven't started this yet, but it is something we need to do. When we plot Veff vs Generation, Theta vs Generation, and R vs Generation we need to add in the value of these for the current ARA bicone as a baseline for our evolutions.
Note: All of the contents over the last month need to be added to the manual! DO NOT FORGET! |
|
|
24
|
Mon Dec 2 16:41:35 2019 |
Julie Rolla | Requesting an interactive job | Requesting an interactive job:
qsub -I -X -l walltime=1:00:00 -l nodes=1:ppn=40
(ppn=24 makes it quicker FYI)
Note that walltime is the length of time you'll need it. You will likely need it for 4+ hours if you are running the loop... Probably longer. Note that before you enter this command into your command line, you must be logged into OSC.
For submitting a job with GPUs (which we will be doing to speed up XF now as of 1/24/20), use this command:
qsub -I -X -l walltime=1:00:00 -l nodes=1:ppn=40:gpus=1:default OUTDATED
srun -A PAS0654 -t 1:00:00 -N 1 -n 40 CURRENT |
|
|
22
|
Sat Nov 30 22:04:30 2019 |
Julie Rolla | GENETIS Loop Work: Fixing Consol Errors | GENETIS Loop Work: Fixing Consol Errors By Julie Rolla
1. Python programs are not running: I think either (1) the user isn't module loading python, or (2) they are loading the wrong version of Python (remember there's a discrepancy for Python2, and Python3).
Solution: I added a line "module load python/3.6-conda5.2" in our araenv.sh script to address the issue. All of the users are sourcing the araenv.sh script in their .bashrc when they login. It's in the directory: /fs/project/PAS0654/BiconeEvolutionOSC. I also added a line in the XF_Loop.sh script (at the top in the "Initialization of Variables" section) that sources araenv.sh just in case the user hasn't put that in their .bashrc. The line reads: source /fs/project/PAS0654/BiconeEvolutionOSC/araenv.sh
Error message fixed:
0
Data successfully read. Data successfully written.
Fitness scores successfully written.
Fitness function concluded.
rm: cannot remove 'runData.csv': No such file or directory
Unable to parse the pattern
^CTraceback (most recent call last):
File "LRPlot.py", line 99, in <module>
plotLR(generations, lengths, radii, g.numGens, g.destination)
File "LRPlot.py", line 45, in plotLR
plt.show()
File "/usr/lib64/python2.7/site-packages/matplotlib/pyplot.py", line 143, in show
_show(*args, **kw)
File "/usr/lib64/python2.7/site-packages/matplotlib/backend_bases.py", line 108, in __call__
self.mainloop()
File "/usr/lib64/python2.7/site-packages/matplotlib/backends/backend_gtk.py", line 83, in mainloop
gtk.main()
KeyboardInterrupt
2. rm runData.csv error: The following error was printed to consol
rm: cannot remove 'runData.csv': No such file or directory
I found these line in section (E) of the loop.
cd "$WorkingDir"
rm runData.csv
python gensData.py 0
cd Antenna_Performance_Metric
python LRPlot.py "$WorkingDir" "$WorkingDir"/Run_Outputs/$RunName 1 $NPOP
cd ..
# Note: gensData.py floats around in the main dir until it is moved to
I think this error is fine. This is how it works: (Step 1) removes runData.csv, if it exists. (Step 2) Runs gensData.py, which creates a new runData.csv. We wanted to delete it before the .py program was run at the start of the run, because gensData.py appends data to runData.csv, if it already exists. (alternatively if runData.csv doesn't exists it creates it-- which is what we want in this case). That's also why we don't delete it later in the "looping part" of XF_Scripts.sh (part G). We want it to append at this point, so we only delete it in the very beginning to make sure it doesn't append to an old run.
3. cp: cannot stat 'Antenna_Performance_Metric/1.uan': No such file or directory (similarly for 2.uan, 3.uan...etc): This error exists just as the fitness score is being calculated -- ie part E. Here is the print:
cp: cannot stat 'Antenna_Performance_Metric/1.uan': No such file or directory
cp: cannot stat 'Antenna_Performance_Metric/2.uan': No such file or directory
Congrats on getting a fitness score!
File "FScorePlot.py", line 1
-*- coding: utf-8 -*-
^
IndentationError: unexpected indent
Note that we have a few errors here. Right now I am directly addressing the error with the "cp" command. It looks like these files are actually being saved as gen_individual#.uan. For example 0_1.uan. The current line in part E says:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/$i.uan "$WorkingDir"/Run_Outputs/$RunName/${i}.uan
done
However, this is not how these files are being saved. I corrected the first iteration of the loop (ie Part E for generation 0) by editing the line to say:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/0_$i.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}.uan
done
And for the looping part of XF_Loop.sh (ie part G) it has been edited to say:
for i in `seq 1 $NPOP`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${gen}_${i}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}.uan
done
4. Made Number of Neutrinos thrown (NNU) in AraSim's setup.txt file a variable: I've added the following line in part D -- as well as the looping section in part G.
These lines replace the number of neutrinos thrown in our setup.txt AraSim file with what ever number you assigned NNT at the top of XF_Loop.sh in the "variables" section. "setup_dummy.txt" is a copy of setup.txt that has NNU=num_nnu instead of a number. NNU is the number of neutrinos thrown (as seen in setup.txt). This "sed" line finds every instance of num_nnu in setup_dummy.txt and replaces it with $NNT (the number you assigned NNT in the variable section at the top of XF_Loop.sh). It then pipes this information into setup.txt (and overwrites the last setup.txt file allowing the number of neutrinos thrown to be as variable changed at the top of this script instead of manually changing it in setup.txt each time. Command works the following way: sed "s/oldword/newwordReplacingOldword/" path/to/filewiththisword.txt > path/to/fileWeAreOverwriting.txt. Both setup.txt and setup_dummy.txt can be found in the directory: /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/
cd "$AraSimExec"
for i in `seq 1 $NPOP`
do
#This next line replaces the number of neutrinos thrown in our setup.txt AraSim file with what ever number you assigned NNT at the top of this program. setup_dummy.txt is a copy of setup.txt that has \
NNU=num_nnu (NNU is the number of neutrinos thrown. This line finds every instance of num_nnu in setup_dummy.txt and replaces it with $NNT (the number you assigned NNT above). It then pipes this infor\
mation into setup.txt (and overwrites the last setup.txt file allowing the number of neutrinos thrown to be as variable changed at the top of this script instead of manually changing it in setup.txt e\
ach time. Command works the following way: sed "s/oldword/newwordReplacingOldword/" path/to/filewiththisword.txt > path/to/fileWeAreOverwriting.txt
sed "s/num_nnu/$NNT/" /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/setup_dummy.txt > /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/setup.txt
#We will want to call a job here to do what this AraSim call is doing so it can run in parallel
cd $WorkingDir
qsub -v num=$i AraSimCall.sh
rm outputs/*.root
done
5. rm cannot remove 'simulation_PEC.xmacro': No such file or directory error appearing in generations after the first (ie not seen in gen 0, but seen in gen 1+).
Statistics initialized.
Tournament parents selected.
Tournament complete.
rm: cannot remove 'simulation_PEC.xmacro': No such file or directory
I found that it tries to remove it twice. See here:
######## Loop (G) ######################################################################################
# 1. Does steps A-F for each generation
###########################################################################################################
for gen in `seq 1 $TotalGens`
do
# used for fixed number of generations
#gen=0; while [ `cat highfive.txt` -eq 0 ]; do (( gen++ ))
# use for runs until convergence
#should the line below be double indented?
read -p "Starting generation ${gen}. Press any key to continue... " -n1 -s
#Part A
cd $XmacrosDir
rm simulation_PEC.xmacro
cd "$WorkingDir"
./roulette_algorithm.exe cont $NPOP
cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv
##chmod -R 777 $WorkingDir
#we can make a function that does this with the flag $gen
#doing this in part A would mean setting $gen=0
#Part B
# First, remove the old .xmacro files
#when do that, we end up making the files only readable; we should just overwrite them
#alternatively, we can just set them as rwe when the script makes them
cd $XmacrosDir
#I'm commenting out the next two lines. They are written to below (around lines 154-170)
#If we keep them this way then the files have restricted permissions
rm output.xmacro
rm simulation_PEC.xmacro
I have commented the first removal of simulation_PEC.xmacro. If there are any issues with it running, we can go ahead and comment it back in; however, I suspect this is not a necessary line to have added in. A run has not been performed since these edits. So, once it has been tested, we can delete the comments out:
#Part A
#I think these next two lines can be deleted. Its repeated again just below...11/30/19 comment by Julie #cd $XmacrosDir #rm simulation_PEC.xmacro
cd "$WorkingDir"
./roulette_algorithm.exe cont $NPOP
cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv
6. Python Error from FScorePlot.py: The error can be seen in the line below.
Congrats on getting a fitness score!
File "FScorePlot.py", line 1
-*- coding: utf-8 -*-
^
IndentationError: unexpected indent
Congrats on getting some nice plots!
I've commented out this strange line at the start of this python program: -*- coding: utf-8 -*-. This fixed the immediate error, but threw another one when I tried to run it outside of the loop with the command
python FScorePlot.py /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie_11_30/ /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie_11_30/ 2
The next error it threw can be seen below.
Traceback (most recent call last):
File "FScorePlot.py", line 79, in <module>
fScores.append(fScorei[2:])
AttributeError: 'numpy.ndarray' object has no attribute 'append'
This is Alex P. I edited the append() and changed it to np.append(fScores, fScorei[2:]) on line 79 in FScorePlot.py
7. rm: cannot remove 'outputs/*.root': No such file or directory: This error comes up right after the AraSim job is submitted in part D.
1011007.pitzer-batch.ten.osc.edu
rm: cannot remove 'outputs/*.root': No such file or directory
Waiting for AraSim jobs to finish...
It's definitely coming from the lines:
for i in `seq 1 $NPOP`
do #We will want to call a job here to do what this AraSim call is doing so it can run in parallel
cd $WorkingDir
qsub -v num=$i AraSimCall.sh rm outputs/*.rootdone
We are currently in $WorkingDir at this point-- which is: WorkingDir= /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop. There is not directory called "outputs" in this location. This is why we are getting an error.Can someone explain what .root files we are trying to remove, and why? I cannot proceed unless someone makes this clear to me, as I do not know what files we are looking to remove (and where they exist/when they are created).
Immediately after that, I get the following error:
Waiting for AraSim jobs to finish...
rm: remove write-protected regular file 'AraSimFlags/1.txt'? y
rm: remove write-protected regular file 'AraSimFlags/2.txt'? y
mv: cannot stat '*.root': No such file or directory
It looks like it is coming from the following line:
cd "$WorkingDir"/Antenna_Performance_Metric
mv *.root "$WorkingDir/Run_Outputs/$RunName/RootFilesGen0/"
However-- again-- it looks like it's trying to move .root files from the directory: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric and put them into: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/$RunName/RootFilesGen0/
It can't move these, because they do not exist where it is trying to take them from. Similarly, the question in bold above would help me with this a lot. From what I can see, we are only utilizing an .txt file output from Arasim. Does Arasim output this to .txt, or are we somehow converting from a .root to a .txt?
This error has not yet been corrected!!!!
Cade's comment here:
The .root files live in BiconeEvolutionOSC/AraSim/outputs/ folder. Thus, we were looking at the wrong directory. They were being removed because it is a giant file (GB size) and are created everytime AraSim runs. It just overwrites itself everytime though so it doesnt add up too much. We can have them deleted later if needed. It's our choice on whether we take it out completely, or redirect to delete the .root files in the correct directory.
8. New errors found after running with these corrections: These need to be fixed!
cp: cannot stat ‘Antenna_Performance_Metric/0_1.uan’: No such file or directory
cp: cannot stat ‘Antenna_Performance_Metric/0_2.uan’: No such file or directory
Congrats on getting a fitness score!
Traceback (most recent call last):
File "FScorePlot.py", line 86, in <module>
Gen = np.zeros(fScores.shape[0]*fScores.shape[1])
I found the cp error. It looks like I was incorrect in #3 about this cp command for the .uan files. I moved all of the currently existing .uan files into a new directory in
/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric/UAN
With all of them out of the Antenna_Performance_Metric directory, I did a run from scratch to see what it creates. Here's the outcome: So on generation 1, for individual one its going
1_1.uan
1_2.uan
1_3.uan
ie-- > individual_frequencyStep.uan
So once it hits all 60 frequencies for individual 1, it does all 60 for individual 2 by saying:
2_1.uan
2_2.uan
2_3.uan
...etc.This isn't great because it never actually cares about generation number. In section E (so generation 0) we currently have it as:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/0_$i.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}.uan
done
and in section G of the looping part (gens 1+) we have:
for i in `seq 1 $NPOP`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${gen}_${i}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}.uan
done
This doesn't seem like it's being output from XF the way we thought it is. I think the easiest way to do this is to edit the xmacros to have it output like:
generation_individual_frequency.uan
and thus
cp Antenna_Performance_Metric/${gen}_${i}_${frequency}.uan
If so, we'd have to make a new variable in the bash script for $frequency to go from 1-60, and have it loop moving all of those. I will be looking into this with Cade tomorrow 12/3/19 (since he is the xmacro script expert). Stay tuned!
Phew....thanks for sticking with me on this long update. |
|