| ID |
Date |
Author |
Subject |
|
230
|
Thu Jul 6 14:52:09 2023 |
Jack Tillman | Building - Matching Circuit Schematic, PCB, and components |
Attached are images of the matching circuit schematic and PCB design. A parts list is also attached in .pdf and .csv format. The .csv format can be imported into Digikey if necessary.
Table of component values:
| Inductors |
Capacitors |
SMA Connectors |
| 22 nH |
7.5 pF |
50 Ω |
| 27 nH |
5.7 pF |
50 Ω |
|
| Attachment 1: Matching_Circuit_Schematic.png
|
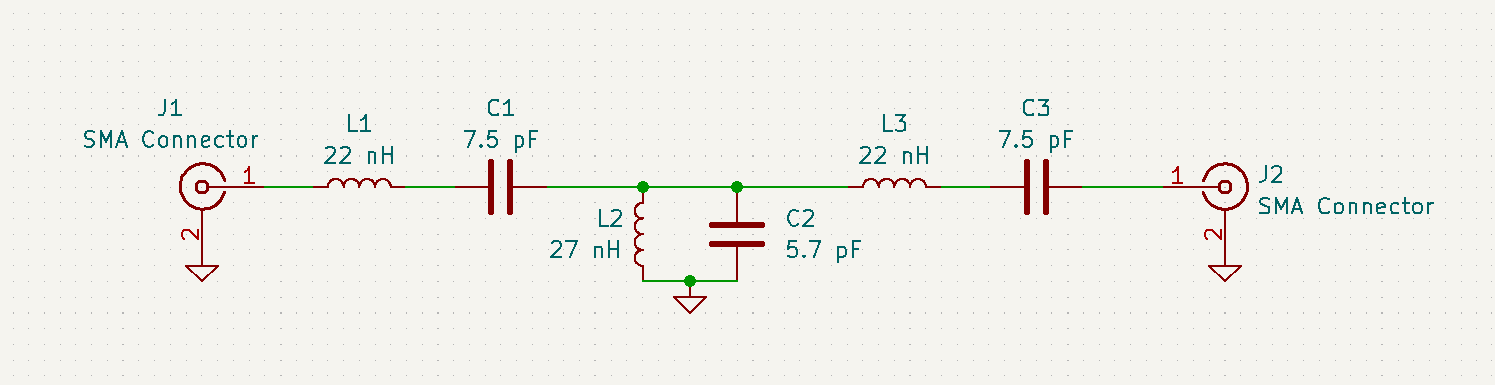 |
| Attachment 2: Matching_Circuit_PCB.png
|
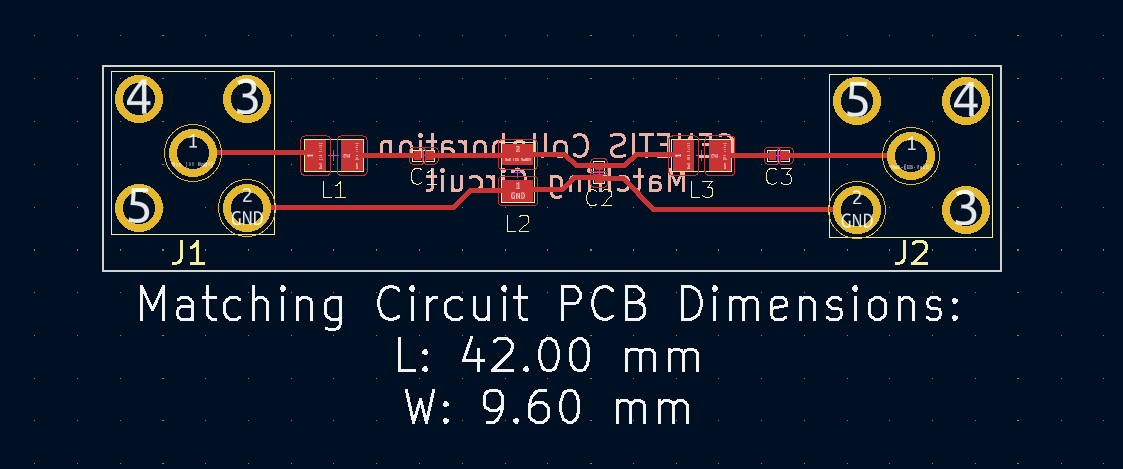 |
| Attachment 3: Matching_Circuit_DigiKey_Cart.pdf
|
| Attachment 4: Matching_Circuit_DigiKey_Cart.csv
|
Index,Quantity,Part Number,Manufacturer Part Number,Description,Available,Backorder,Unit Price,Extended Price USD
1,3,WM26450-ND,733910083,"SMA RA JACK, PCB",3,0,5.1,15.3
2,4,490-GJM0335C1E7R5BB01DCT-ND,GJM0335C1E7R5BB01D,CAP CER 7.5PF 25V C0G/NP0 0201,4,0,0.1,0.4
3,2,490-17198-1-ND,GJM0335C1H5R7CB01D,CAP CER 5.7PF 50V C0G/NP0 0201,2,0,0.09,0.18
4,2,490-16032-1-ND,LQW2BAN27NJ00L,FIXED IND 27NH 2A 70 MOHM SMD,2,0,0.42,0.84
5,4,490-16028-1-ND,LQW2BAN22NJ00L,FIXED IND 22NH 1.9A 70 MOHM SMD,4,0,0.42,1.68
|
|
111
|
Mon Oct 5 21:06:29 2020 |
Everyone | Data Runs |
Machtay_20200831_Asym_Length_and_Angle 10 individuals
Machtay_20200911_Symmetric 10 individuals, fewer neutrinos
Machtay_20200914_Asymmetric_50_Individuals 50 individuals, fewer neutrinos
Machtay_20200929_Asymmetric_test_2 50 individuals, fewer neutrinos, broaden parameter range
|
|
109
|
Fri Oct 2 17:11:25 2020 |
Ethan Fahimi | Update Friday 10/2/2020 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
Today I started working on improving the plots. I started with plotting the average fitness score for each generation on top of all the individuals data points. I made the average fitness score a star with higher contrast so it will stand out. Here is the plot of the first 26 generations of the "Machtay_20200914_Asymmetric_50_Individuals" run with the new averages. |
On monday I'm going to try and create the plot of 2 runs overlaid on top of eachother as Kai suggested. |
| Evelyn |
|
|
| Ryan |
Worked with Ben and Alex P. on making the Bicone function modular on our copy. We finished writing a new roulette selection function, tournament selection function, and reproduction function. |
Finish writing the mutation and crossover functions. After these are complete apply these functions in place of the existing function that is wrong and test to make sure the results are not producing errors. |
| Ben |
|
|
| Ethan |
Updated the AREA code with Alex M's help to work using SLURM. Began a trial run to see if our updates work. |
Begin making changes that allow the job to run faster as well as investigate what is causing some individuals to have fitness scores beyond what should be physically possible. |
|
| Attachment 1: Screen_Shot_2020-10-02_at_5.39.33_PM.png
|
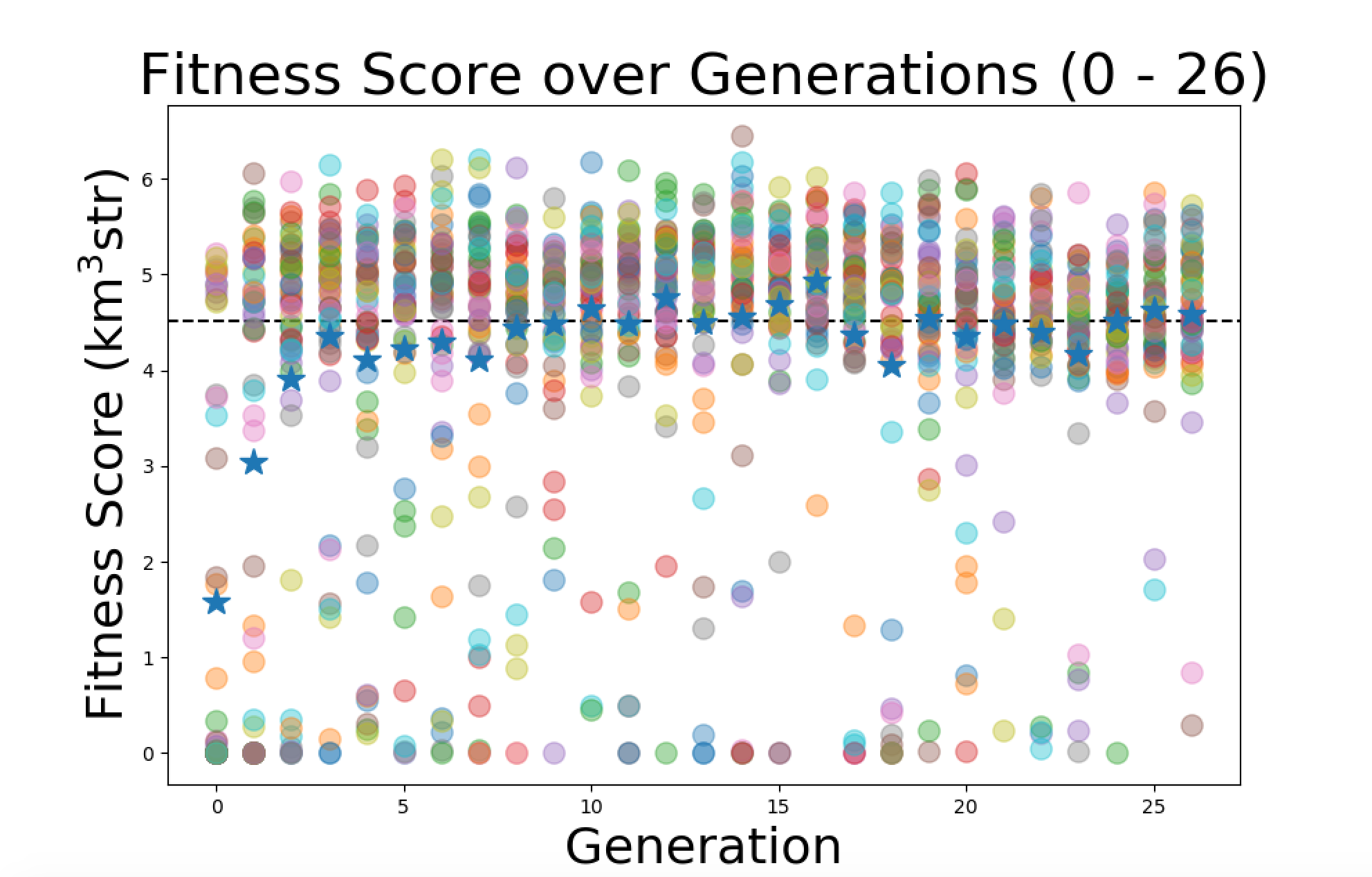 |
|
115
|
Mon Oct 19 17:02:12 2020 |
Ethan Fahimi | Daily Update 10/19 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Alex P. and I put the outer radius constraint into the Asymmetric our version of the algorithm. I have also created the pseudo-fitness function to be able to do some optimization testing that bypasses the time-consuming parts of the run. All I need to do to finish the pseudo tests is to create a loop to run through the generations and plotting procedures. |
|
| Ben |
|
|
| Ethan |
Tried to fix permission issues, as well as work on running the loop (invalid id) |
Continue fixing AREA permission issues with Ben's help. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
116
|
Mon Oct 26 17:59:10 2020 |
Ethan Fahimi | Daily Update 10/26 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
I have the testing loop finished with plotted results now. The program was able to reach optimal results very quickly. It used 100% tournament selection with a cutoff on the outer radius. The algorithm was using an asymmetric algorithm and the ideal bicone it was being compared to was an arbitrarily picked one from an actual symmetric run individual we knew to stay within the outer radius. All individuals for each generation are plotted on this graph. And the fact that these bicones started as asymmetric shows that we can very easily find symmetric answers if they are indeed ideal. |
|
| Ben |
|
|
| Ethan |
Tried to fix permission issues with AREA, as well as learned about running the loop and listening to Jorge's thoughts on our project. |
Possibly move the AREA project onto the project space as it may solve permissions issues. Alex is looking into it. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
| Attachment 1: fitness.png
|
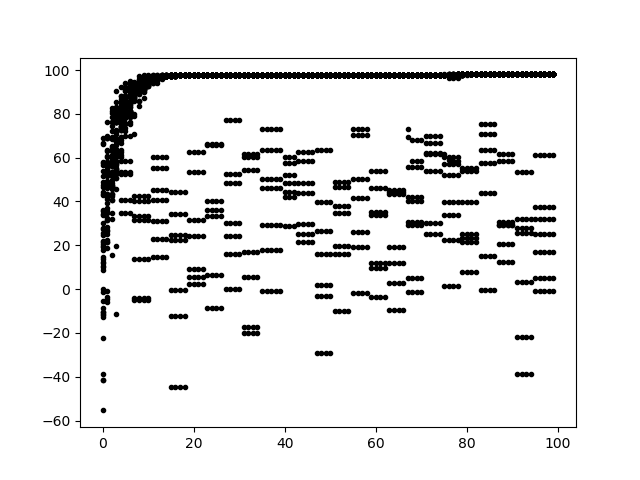 |
|
117
|
Fri Oct 30 17:30:16 2020 |
Ethan Fahimi | Daily Update 10/30 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Attempted to make a new mutation function for the algorithm to try and address the concerns about hitting local maximums from Wednesday. Unfortunately, the idea was unsuccessful when I put it through testing. I would like some input from some of the experts before trying something else. Otherwise, the version I had earlier this week has still been very consistent about optimizing the runs outside of sometimes hitting a local max at about 90/100.
|
|
| Ben |
|
|
| Ethan |
Moved AREA onto my own directory with Alex's help. Began fixing issues with it (small win, no more permissions issues!). |
Continue fixing issues with AraSim on my user. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
Draft
|
Mon Nov 30 16:59:39 2020 |
Ethan Fahimi | Daily Update 10/30 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Attempted to make a new mutation function for the algorithm to try and address the concerns about hitting local maximums from Wednesday. Unfortunately, the idea was unsuccessful when I put it through testing. I would like some input from some of the experts before trying something else. Otherwise, the version I had earlier this week has still been very consistent about optimizing the runs outside of sometimes hitting a local max at about 90/100.
|
|
| Ben |
|
|
| Ethan |
Moved AREA onto my own directory with Alex's help. Began fixing issues with it (small win, no more permissions issues!). |
Continue fixing issues with AraSim on my user. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
122
|
Mon Nov 30 17:00:31 2020 |
Ethan Fahimi | Monday Updates |
| Alex M |
Kicked the loop back up. Helped Ethan and Parker with their projects during the working meeting. |
| Ryan |
Changed the tournament/roulette ratio, reproduction_no, and crossover_no to be read in variables in the GA to increase the quality of life when running the algorithm and to prep the code for some testing. Format for how to call the code is written at the top of the cpp program file. |
| Ethan F |
Worked with Alex M on further fixing AREA. The first generation works now, we are making minor fixes to get subsequent generations up and running. |
| |
|
| |
|
| |
|
| |
|
|
|
125
|
Fri Dec 11 17:47:48 2020 |
Ethan Fahimi | Friday Updates |
| Ethan F |
Fixed the issued AREA was having with finding test_{ind}.txt, now to fix problems with finding Veff and the project should be working. |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
127
|
Tue Jan 26 17:17:41 2021 |
Ethan Fahimi | Tuesday Updates 01/26/2021 |
| Ethan F |
Continued implementing some solutions the OSC helpdesk gave me for fixing the unwanted extensions on the job output files. |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
133
|
Wed Jun 23 16:34:10 2021 |
Ethan Fahimi | Daily Update 6/23/2021 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Lydon |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
With Alex M's help, managed to get AREA working, plotted results. |
The results look relatively flat, possibly the GA is unoptimized, may need work. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
134
|
Wed Jul 14 15:45:30 2021 |
Ethan Fahimi | Wednesday Updates (7/14/2021) |
| Ethan |
Worked with Alex on fixing a few bugs with AREA. We are trying to solve an issue where the individuals are not finishing runs (around one in every four gens with 100 individuals). We believe some individuals may be too good and are then taking more than the wall time we have given them. Alex is testing this while I am working on a script that will add all the weights in the temp_{ind}.txt files. (See weightAdder.py for more) |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
138
|
Fri Sep 17 13:41:36 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot |
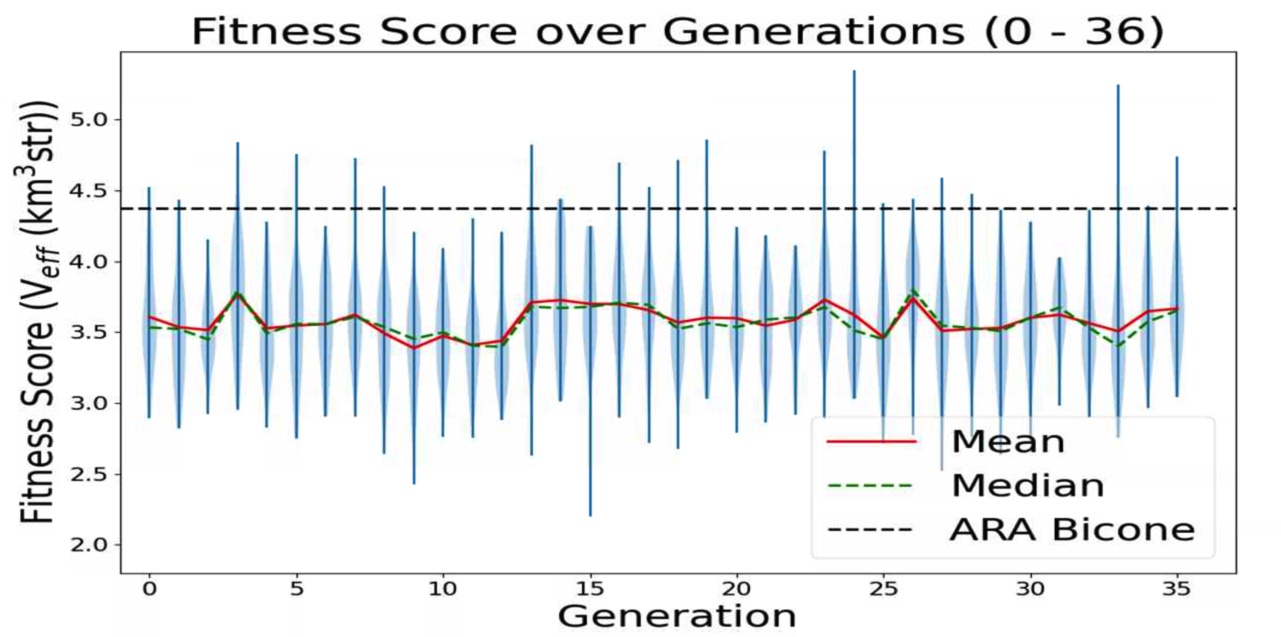
This is a plot made from the AREA project with full Arasim implementation. It can be seen that the Veff of any individuals is not what I would consider "good", nor is it really rising, it is quite flat. This is because in this version of AREA, the gain pattern at each frequency is generated differently than each other frequency, there is no correlation. This is known and actively being corrected. This plot is of old data and was just made for two reasons: to make sure that the violin plotting script works for AREA, to display this early form of AREA that has been adapted for full Arasim.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
This plot is further detailed in Julie Rolla's doctorate thesis. |
| Attachment 1: Image_9-13-21_at_6.35_PM.jpg
|
 |
|
Draft
|
Mon Nov 8 17:04:30 2021 |
Ethan Fahimi | 11/04/2021 AREA run 2 violin plot |
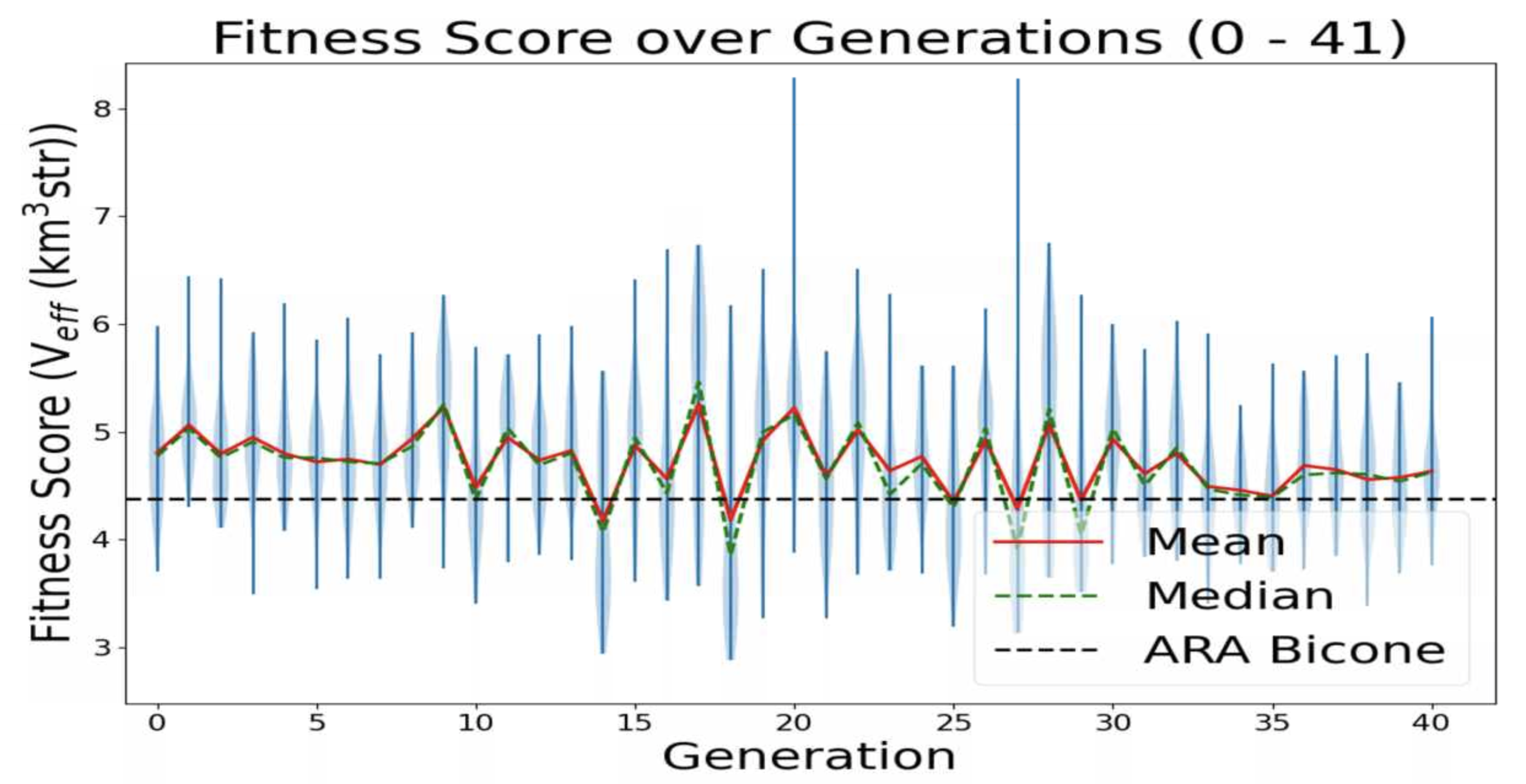
This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation. |
| Attachment 1: 20211104fahimi5run2.png
|
 |
|
140
|
Mon Nov 8 17:27:01 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot |
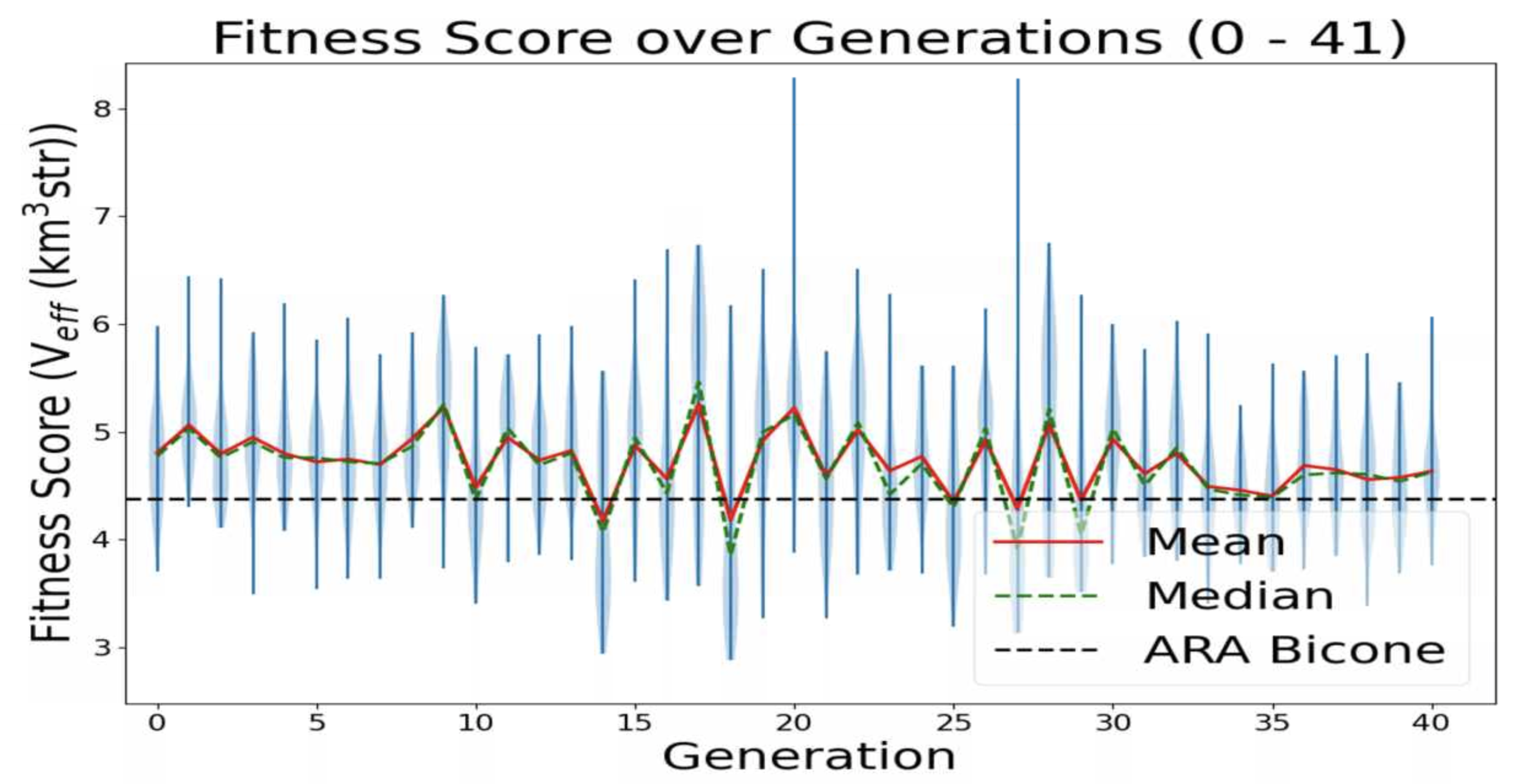
This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
individual 32 in gen 20 and individual 35 in gen 27 look promising (they have Veff > 8) |
| Attachment 1: 20211104fahimi5run2.png
|
 |
|
161
|
Tue Jun 7 14:14:21 2022 |
Dylan Wells | Matching Circuits Slides |
Slides contatining my notes on matching circuits.
https://docs.google.com/presentation/d/1x25nhiqaW7LvPZ1pNZ5O4ZzsWZbtgqxBQ5haB9uWgQY/edit?usp=sharing |
|
170
|
Tue Jun 28 13:27:13 2022 |
Dylan Wells | Changes needed for the matching circuit script |
-
Fix the functions for the SLPC, SCPL, and PLSC L networks (change the paramaters to match with the format of our data)
-
Write the PCSL function
-
Create a function to find the number of L networks necessary (N) given a source and load resistance as well as a frequency range.
-
Write a function to broadband match two impedances given a source, load, central frequency, and N. (return a list of capacitances and inductances for the L networks)
|
|
178
|
Wed Aug 10 22:38:20 2022 |
Dylan Wells | Instructions for Installing IceMC |
Installing IceMC:
I: Getting anitaBuildTool
Clone the anitaBuildTool repository (https://github.com/anitaNeutrino/anitaBuildTool) to your user.
II: Get the Anita.sh file onto your user.
Either copy the file from /users/PAS1960/dylanwells1629/Anita.sh
or create a new file Anita.sh with the following code
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
#this modules were originally loading in both the env.sh, and bashrc_anita.sh files. This was redundant so it was added here, and removed from the others.
module load gnu/7.3.0
module load gnu
module load mvapich2
module load fftw3
#module load python/3.6-conda5.2
module load cmake
PATH=$PATH:$HOME/.local/bin:$home/bin
export PATH
export CC=`which gcc`
export CXX=`which g++`
export FFTWDIR=/fs/project/PAS0654/shared_software/fftw3/gnu/6.3/mvapich2/2.2/3.3.5
export ANITA_SOURCE_DIR=~/anitaBuildTool/
export ANITA_UTIL_INSTALL_DIR=~/anitaBuildTool/
export ICEMC_SRC_DIR=~/anitaBuildTool/components/icemc/
export ICEMC_BUILD_DIR=~/anitaBuildTool/build/components/icemc/
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
# User specific aliases and functions
#This env.sh is for running the BiconeEvolution GENETIS software. This should only be un-commented if you are running GENETIS software. When you do this, comment out env.sh.
#source ~/new_root_setup.sh
source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh
#source /cvmfs/ara.opensciencegrid.org/trunk/centos7/setup.sh
#module load python/3.6-conda5.2
#BiconeGENETIS directory shortcut SHARED
alias GE='cd ../../../fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/'
#emacs Alias
alias emacs='emacs -nw'
#root alias
alias root='root -l'
#Alias
alias l="ls"
alias python='/cvmfs/ara.opensciencegrid.org/trunk/centos7/misc_build/bin/python3.9'
Then source the file
Note: You will need access to PAS0654 for this step or you will get a permissions error.
III: Running the build tool.
Go into the anitaBuildTool directory
And run the building script
Note: There will be an error if you source files for running Ara in your .bashrc
Comment these out and restart your terminal before running the build. (remember to source Anita.sh before running the build tool. You could also source Anita.sh in your .bashrc)

Error if you source files for running Ara:
CMake Error at components/libRootFftwWrapper/cmake_install.cmake:238 (file):
file INSTALL cannot copy file
"/users/PAS1960/dylanwells1629/anitaBuildTool/components/libRootFftwWrapper/include/AnalyticSignal.h"
to
"/cvmfs/ara.opensciencegrid.org/v2.0.0/centos7/ara_build/include/AnalyticSignal.h":
Read-only file system.
Call Stack (most recent call first):
|
|
179
|
Tue Aug 16 11:41:07 2022 |
Dylan Wells | Instructions for Running IceMC |
Running IceMC:
Go into the directory ../anitaBuildTool/build/components/icemc/
And run the command
*May need to chmod -R 775 ../anitaBuildTool/comonents/icemc/ if you get a permissions error
inputFile:
Must be the full path to the file
Config files are found in ../anitaBuildTool/components/icemc
Ex: /users/PAS1960/dylanwells1629/anitaBuildTool/components/icemc/inputs.anita4.conf
Config files are found in ../anitaBuildTool/components/icemc
outputDirectory:
Will be made in ../anitaBuildTool/build/components/icemc/ by default, specify full path otherwise.
runNumber:
The run number.
numberOfNeutrinos:
The number of neutrinos generated in the simulation.
Can be found in inputs.conf
Default is 2,000,000.
#How many neutrinos to generate
triggerThreshold:
Threshold for each band for the trigger.
Default is 2.3
#thresholds for each band- this is only for the frequency domain voltage trigger. If using a different trigger scheme then keep these at the default values of 2.3 because the max among them is used for the chance in hell cuts
energyExponent:
The exponent of the energy for the neutrinos
Can be found in input.conf
Default is 1020
# Select energy (just enter the exponent) or (30) for baseline ES&S (1) for E^-1 (2) for E^-2 (3) for E^-3 (4) for E^-4 (5) for ES&S flux with cosmological constant (6) for neutrino GZK flux from Iron nuclei (16-22)not using spectrum but just for a single energy (101-114)use all kinds of theoretical flux models |
|
181
|
Thu Aug 18 13:03:47 2022 |
Dylan Wells | Comparing inputs and outputs between AraSim and IceMC |
Comparing Inputs:
Frequency Lists:
Ara - 83.33MHz - 1066.70 MHz, step = 13.33MHz
IceMC - 200MHz - 1500MHz, step = 10MHz
Number of files read in:
Ara - 1
IceMC - 8
Formating of files:
Ara - Theta, Phi, Gain (dB, thetra), Gain (theta), Phase (theta)
IceMc - Frequency, Gain (dB) (Different files cover different thetas and phis)
File Type:
Ara - .txt
IceMC - no suffix (file with 2 columns of text)
Other notes for IceMC inputs:
IceMC reads in 8 different files for gain.
vv_0 hh_0 vh_0 hv_0 vv_el vv_az hh_el hh_az
Found in ../anitaBuiltTool/components/icemc/data
vv_0 = gains for vertical polarization
hh_0 = gains for horizontal polarization
vh_0 = gains for v → h cross polarization
hv_0 = gains for h → v cross polarization
vv_el = v polarization, e angle
vv_az = v polarization, a angle
hh_el = h polarization, e angle
hh_az = h polarization a angle
for e angle and a angle in
0. 0
-
5
-
10
-
20
-
30
-
45
-
90
(iterates 1 to 6)
Comparing Outputs:
IceMC - veff is in second column in veff+runName+.txt file (in the outputDirectory directory)
Ara - veff is at the bottom of the AraOut.txt file |