| ID |
Date |
Author |
Subject |
Project |
|
5
|
Tue Apr 18 12:02:55 2017 |
Amy Connolly | How to ship to Pole | Hardware | Here is an old email thread about how to ship a station to Pole.
|
| Attachment 1: Shipping_stuff_to_Pole__a_short_how_to_from_you_would_be_nice_.pdf
|
| Attachment 2: ARA_12-13_UH_to_CHC_Packing_List_Box_2_of_3.pdf
|
| Attachment 3: ARA_12-13_UH_to_CHC_Packing_List_Box_1_of_3.pdf
|
| Attachment 4: ARA_12-13_UH_to_CHC_Packing_List_Box_3_of_3.pdf
|
| Attachment 5: IMG_4441.jpg
|  |
|
12
|
Fri Aug 25 12:34:47 2017 |
Amy Connolly - posting stuff from Todd Thompson | How to run coffee | Other | One really useful thing in here is how to describe "Value Added" to visitors. |
| Attachment 1: guidelines.pdf
|
|
49
|
Thu Sep 14 22:30:06 2023 |
Jason Yao | How to profile a C++ program | Software | This guide is modified from section (d) of the worksheet inside Module 10 of Phys 6810 Computational Physics (Spring 2023).
NOTE: gprof does not work on macOS. Please use a linux machine (such as OSC)
To use gprof, compile and link the relevant codes with the -pg option:
Take a look at the Makefile make_hello_world and modify both the CFLAGS and LDFLAGS lines to include -pg
Compile and link the script by typing
make -f make_hello_world
Execute the program
./hello_world.x
With the -pg flags, the execution will generate a file called gmon.out that is used by gprof.
The program has to exit normally (e.g. we can't stop with a ctrl-C).
Warning: Any existing gmon.out file will be overwritten.
Run gprof and save the output to a file (e.g., gprof.out) by
gprof hello_world.x > gprof.out
We should at this point see a text file called gprof.out which contains the profile of hello_world.cpp
vim gprof.out |
| Attachment 1: hello_world.cpp
|
#include <iostream>
#include <thread>
#include <chrono>
#include <cmath>
using namespace std;
void nap(){
// usleep(3000);
// this_thread::sleep_for(30000ms);
for (int i=0;i<1000000000;i++){
double j = sqrt(i^2);
}
}
int main(){
cout << "taking a nap" << endl;
nap();
cout << "hello world" << endl;
}
|
| Attachment 2: make_hello_world
|
SHELL=/bin/sh
# Note: Comments start with #. $(FOOBAR) means: evaluate the variable
# defined by FOOBAR= (something).
# This file contains a set of rules used by the "make" command.
# This makefile $(MAKEFILE) tells "make" how the executable $(COMMAND)
# should be create from the source files $(SRCS) and the header files
# $(HDRS) via the object files $(OBJS); type the command:
# "make -f make_program"
# where make_program should be replaced by the name of the makefile.
#
# Programmer: Dick Furnstahl (furnstahl.1@osu.edu)
# Latest revision: 12-Jan-2016
#
# Notes:
# * If you are ok with the default options for compiling and linking, you
# only need to change the entries in section 1.
#
# * Defining BASE determines the name for the makefile (prepend "make_"),
# executable (append ".x"), zip archive (append ".zip") and gzipped
# tar file (append ".tar.gz").
#
# * To remove the executable and object files, type the command:
# "make -f $(MAKEFILE) clean"
#
# * To create a zip archive with name $(BASE).zip containing this
# makefile and the SRCS and HDRS files, type the command:
# "make -f $(MAKEFILE) zip"
#
# * To create a gzipped tar file with name $(BASE).tar.gz containing this
# makefile and the source and header files, type the command:
# "make -f $(MAKEFILE) tarz"
#
# * Continuation lines are indicated by \ with no space after it.
# If you get a "missing separator" error, it is probably because there
# is a space after a \ somewhere.
#
###########################################################################
# 1. Specify base name, source files, header files, input files
###########################################################################
# The base for the names of the makefile, executable command, etc.
BASE= hello_world
# Put all C++ (or other) source files here. NO SPACES after continuation \'s.
SRCS= \
hello_world.cpp
# Put all header files here. NO SPACES after continuation \'s.
HDRS= \
# Put any input files you want to be saved in tarballs (e.g., sample files).
INPFILE= \
###########################################################################
# 2. Generate names for object files, makefile, command to execute, tar file
###########################################################################
# *** YOU should not edit these lines unless to change naming conventions ***
OBJS= $(addsuffix .o, $(basename $(SRCS)))
MAKEFILE= make_$(BASE)
COMMAND= $(BASE).x
TARFILE= $(BASE).tar.gz
ZIPFILE= $(BASE).zip
###########################################################################
# 3. Commands and options for different compilers
###########################################################################
#
# Compiler parameters
#
# CXX Name of the C++ compiler to use
# CFLAGS Flags to the C++ compiler
# CWARNS Warning options for C++ compiler
# F90 Name of the fortran compiler to use (if relevant)
# FFLAGS Flags to the fortran compiler
# LDFLAGS Flags to the loader
# LIBS A list of libraries
#
CXX= g++
CFLAGS= -g
CWARNS= -Wall -W -Wshadow -fno-common
MOREFLAGS= -Wpedantic -Wpointer-arith -Wcast-qual -Wcast-align \
-Wwrite-strings -fshort-enums
# add relevant libraries and link options
LIBS=
# LDFLAGS= -lgsl -lgslcblas
LDFLAGS=
###########################################################################
# 4. Instructions to compile and link, with dependencies
###########################################################################
all: $(COMMAND)
.SUFFIXES:
.SUFFIXES: .o .mod .f90 .f .cpp
#%.o: %.mod
# This is the command to link all of the object files together.
# For fortran, replace CXX by F90.
$(COMMAND): $(OBJS) $(MAKEFILE)
$(CXX) -o $(COMMAND) $(OBJS) $(LDFLAGS) $(LIBS)
# Command to make object (.o) files from C++ source files (assumed to be .cpp).
# Add $(MOREFLAGS) if you want additional warning options.
%.o: %.cpp $(HDRS) $(MAKEFILE)
$(CXX) -c $(CFLAGS) $(CWARNS) -o $@ $<
# Commands to make object (.o) files from Fortran-90 (or beyond) and
# Fortran-77 source files (.f90 and .f, respectively).
.f90.mod:
$(F90) -c $(F90FLAGS) -o $@ $<
.f90.o:
$(F90) -c $(F90FLAGS) -o $@ $<
.f.o:
$(F90) -c $(FFLAGS) -o $@ $<
##########################################################################
# 5. Additional tasks
##########################################################################
# Delete the program and the object files (and any module files)
clean:
/bin/rm -f $(COMMAND) $(OBJS)
/bin/rm -f $(MODIR)/*.mod
# Pack up the code in a compressed gnu tar file
tarz:
tar cfvz $(TARFILE) $(MAKEFILE) $(SRCS) $(HDRS) $(MODIR) $(INPFILE)
# Pack up the code in a zip archive
zip:
zip -r $(ZIPFILE) $(MAKEFILE) $(SRCS) $(HDRS) $(MODIR) $(INPFILE)
##########################################################################
# That's all, folks!
##########################################################################
|
|
50
|
Wed Jun 12 12:10:05 2024 |
Jacob Weiler | How to install AraSim on OSC | Software | # Installing AraSim on OSC
Readding this because I realized it was deleted when I went looking for it 
Quick Links:
- https://github.com/ara-software/AraSim # AraSim github repo (bottom has installation instructions that are sort of right)
- Once AraSim is downloaded: AraSim/UserGuideTex/AraSimGuide.pdf (manual for AraSim) might have to be downloaded if you can't view pdf's where you write code
Step 1:
We need to add in the dependancies. AraSim needs multiple different packages to be able to run correctly. The easiest way on OSC to get these without a headache is to add the following to you .bashrc for your user.
cvmfs () {
module load gnu/4.8.5
export CC=`which gcc`
export CXX=`which g++`
if [ $# -eq 0 ]; then
local version="trunk"
elif [ $# -eq 1 ]; then
local version=$1
else
echo "cvmfs: takes up to 1 argument, the version to use"
return 1
fi
echo "Loading cvmfs for AraSim"
echo "Using /cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
source "/cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
#export JUPYTER_CONFIG_DIR=$HOME/.jupyter
#export JUPYTER_PATH=$HOME/.local/share/jupyter
#export PYTHONPATH=/users/PAS0654/alansalgo1/.local/bin:/users/PAS0654/alansalgo1/.local/bin/pyrex:$PYTHONPATH
}
If you want to view my bashrc
- /users/PAS1977/jacobweiler/.bashrc
Reload .bashrc
- source ~/.bashrc
Step 2:
Go to directory that you want to put AraSim and type:
- git clone https://github.com/ara-software/AraSim.git
This will download the github repo
Step 3:
We need to use make and load sourcing
- cd AraSim
- cvmfs
- make
wait and it should compile the code
Step 4:
We want to do a test run with 100 neutrinos to make sure that it does *actually* run
Try: - ./AraSim SETUP/setup.txt
This errored for me (probably you as well)
Switch from frequency domain to time domain in the setup.txt
- cd SETUP
- open setup.txt
- scroll to bottom
- Change SIMULATION_MODE = 1
- save
- cd ..
- ./AraSim SETUP/setup.txt
This should run quickly and now you have AraSim setup! |
|
14
|
Mon Sep 18 12:06:01 2017 |
Oindree Banerjee | How to get anitaBuildTool and icemc set up and working | Software | First try reading and following the instructions here
https://u.osu.edu/icemc/new-members-readme/
Then e-mail me at oindreeb@gmail.com with your problems
|
|
24
|
Wed Jun 6 17:48:47 2018 |
Jorge Torres | How to build ROOT 6 on an OSC cluster | | Disclaimer: I wrote this for Owens, which I think will also work on Pitzer. I recommend following Steven's instructions, and use mine if it fails to build. J
1. Submit a batch job so the processing resources are not limited (change the project ID if needed.):
qsub -A PAS0654 -I -l nodes=1:ppn=4,walltime=2:00:00
2. Reset and load the following modules (copy and paste as it is):
module reset
module load cmake/3.7.2
module load python/2.7.latest
module load fftw3/3.3.5
3. Do echo $FFTW3_HOME and make sure it spits out "/usr/local/fftw3/intel/16.0/mvapich2/2.2/3.3.5". If it doesn't, do
export FFTW3_HOME=/usr/local/fftw3/intel/16.0/mvapich2/2.2/3.3.5
Otherwise, just do
export FFTW_DIR=$FFTW3_HOME
4. Do (Change DCMAKE_INSTALL_PREFIX and point it to the root source directory)
cmake -DCMAKE_C_COMPILER=`which gcc` \
-DCMAKE_CXX_COMPILER=`which g++` \
-DCMAKE_INSTALL_PREFIX=${HOME}/local/oakley/ROOT-6.12.06 \
-DBLAS_mkl_intel_LIBRARY=${MKLROOT}/lib/intel64 \
../root-6.12.06 2>&1 | tee cmake.log
It will configure root so it can be installed in the machine (takes about 5 minutes).
5. Once it is configured, do the following to build root (takes about 45 min)
make -j4 2>&1 | tee make.log
6. Once it's done, do
make install
In order to run it, now go into the directory, then cd bin. Once you're in there you should see a .sh called 'thisroot.sh'. Type 'source thisroot.sh'. You should now be able to type 'root' and it will run. Note that you must source this EVERY time you log into OSC. The smart thing to do would be to put this into your bash script.
(Second procedure from S. Prohira)
1. download ROOT: https://root.cern.ch/downloading-root (whatever the latest pro release is)
2. put the source tarball somewhere in your directory on ruby and expand it into the "source" folder
3. on ruby, open your ~/.bashrc file and add the following lines:
export CC="/usr/local/gnu/7.3.0/bin/gcc"
export CXX="/usr/local/gnu/7.3.0/bin/g++"
module load cmake
module load python
module load gnu/7.3.0
4. then run: source ~/.bashrc
5. make a "build" directory somewhere else on ruby called 'root' or 'root_build' and cd into that directory.
6. do: cmake /path/to/source/folder (e.g. the folder you expanded from the .tar file above. should finish with no errors.) here you can also include the -D flags that you want (such as minuit2 for the anita tools)
-for example, the ANITA tools need you to do: cmake -Dminuit2:bool=true /path/to/source/folder.
7. do: make -j4 (or the way that Jorge did it above, if you want to submit it as a batch job (and not be a jerk running a job on the login nodes like i did))
8. add the following line to your .bashrc file (or .profile, whatever startup file you prefer):
source /path/to/root/build/directory/bin/thisroot.sh
9. enjoy root!
|
|
23
|
Wed Jun 6 08:54:44 2018 |
Brian Clark and Oindree Banerjee | How to Access Jacob's ROOT6 on Oakley | | Source the attached env.sh file. Good to go! |
| Attachment 1: env.sh
|
export ROOTSYS=/users/PAS0174/osu8620/root-6.08.06
eval 'source /users/PAS0174/osu8620/root-6.08.06/builddir/bin/thisroot.sh'
export LD_INCLUDE_PATH=/users/PAS0174/osu8620/cint/libcint/build/include:$LD_INCLUDE_PATH
module load fftw3/3.3.5
module load gnu/6.3.0
module load python/3.4.2
module load cmake/3.7.2
#might need this, but probably not
#export CC=/usr/local/gcc/6.3.0/bin/gcc
|
|
19
|
Mon Mar 19 12:27:59 2018 |
Brian Clark | How To Do an ARA Monitoring Report | Other | So, ARA has five stations down in the ice that are taking data. Weekly, a member of the collaboration checks on the detectors to make sure that they are healthy.
This means things like making sure they are triggering at approximately the right rates, are taking cal pulsers, that the box isn't too hot, etc.
Here are some resources to get you started. Usual ara username and password apply in all cases.
Also, the page where all of the plots live is here: http://aware.wipac.wisc.edu/
Thanks, and good luck monitoring! Ask someone whose done it before when in doubt.
Brian |
|
2
|
Thu Mar 16 10:39:15 2017 |
Amy Connolly | How Do I Connect to the ASC VPN Using Cisco and Duo? | | For Mac and Windows:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=14542
For Linux, in case some of your students need it:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=17908
From Sam 01/25/17: It doesn't work from my Ubuntu 14 machine. My VPN setup in 14 does not have the "Software Token Authentication" option on the screen as shown in the instructions. It fails on connection attempt.
The instructions specify Ubuntu 16; perhaps there is a way to make it work on 14, but I don't know what it is.
|
|
37
|
Tue May 14 10:38:08 2019 |
Amy | Getting started with AraSim | Software | Attached is a set of slides on Getting Started with QC, a simulation monitoring project. It has instructions on getting started in using a terminal window, and downloading, compiling and running AraSim, the simulation program for the ARA project. AraSim has moved from the SVN repository to github, and so now you should be able to retrieve it, compile it using:
git clone https://github.com/ara-software/AraSim.git
cd AraSim
make
./AraSim
It will run without arguments, but the output might be silly. You can follow the instructions for running AraSim that are in the qc_Intro instructions, which will give them not silly results. Those parts are still correct.
You might get some const expr errors if you are using ROOT 6, such as the ones in the first screen grab below. As mentioned in the error messages, you need to change from const expr to not. A few examples are shown in the next screen grab.
If you are here, you likely would also want to know how to install the prerequisites themselves. You might find this entry helpful then: http://radiorm.physics.ohio-state.edu/elog/How-To/4. It is only technically applicable to an older version that is designed for compatibility with ROOT5, but it will give you the idea.
These instructions are also superceded by an updated presentation at http://radiorm.physics.ohio-state.edu/elog/How-To/38
|
| Attachment 1: intro_to_qc.pdf
|
| Attachment 2: Screenshot_from_2019-05-14_10-54-48.png
|  |
| Attachment 3: Screenshot_from_2019-05-14_10-56-19.png
|  |
|
21
|
Fri Mar 30 12:06:11 2018 |
Brian Clark | Get icemc running on Kingbee and Unity | Software | So, icemc has some needs (like Mathmore) and preferably root 6 that aren't installed on kingbeen and unity.
Here's what I did to get icecmc running on kingbee.
Throughout, $HOME=/home/clark.2668
- Try to install new version fo ROOT (6.08.06, which is the version Jacob uses on OSC) with CMAKE. Failed because Kingbee version of cmake is too old.
- Downloaded new version of CMAKE (3.11.0), failed because kingbee doesn't have C++11 support.
- Downloaded new version of gcc (7.33) and installed that in $HOME/QCtools/source/gcc-7.3. So I installed it "in place".
- Then, compiled the new version of CMAKE, also in place, so it's in $HOME/QCtools/source/cmake-3.11.0.
- Then, tried to compile ROOT, but it got upset because it couldn't find CXX11; so I added "export CC=$HOME/QCtools/source/gcc-7.3/bin/gcc" and then it could find it.
- Then, tried to compile ROOT, but couldn't because ROOT needs >python 2.7, and kingbee has python 2.6.
- So, downloaded latest bleeding edge version of python 3 (pyton 3.6.5), and installed that with optimiation flags. It's installed in $HOME/QCtools/tools/python-3.6.5-build.
- Tried to compile ROOT, and realized that I need to also compile the shared library files for python. So went back and compiled with --enable-shared as an argument to ./configure.
- Had to set the python binary, include, and library files custom in the CMakeCache.txt file.
|
|
33
|
Mon Feb 11 21:58:26 2019 |
Brian Clark | Get a quick start with icemc on OSC | Software | Follow the instructions in the attached "getting_started_with_anita.pdf" file to download icemc, compile it, generate results, and plot those results. |
| Attachment 1: sample_bash_profile.sh
|
# .bash_profile
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
# User specific environment and startup programs
PATH=$PATH:$HOME/.local/bin:$HOME/bin
export PATH
|
| Attachment 2: sample_bashrc.sh
|
# .bashrc
source bashrc_anita.sh
# we also want to set two more environment variables that ANITA needs
# you should update ICEMC_SRC_DIR and ICEMC_BUILD_DIR to wherever you
# downloaded icemc too
export ICEMC_SRC_DIR=/path/to/icemc #change this line!
export ICEMC_BUILD_DIR=/path/to/icemc #change this line!
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
|
| Attachment 3: test_plot.cc
|
//C++ includes
#include <iostream>
//ROOT includes
#include "TCanvas.h"
#include "TStyle.h"
#include "TH1D.h"
#include "TFile.h"
#include "TTree.h"
using namespace std;
int main(int argc, char *argv[])
{
if(argc<2)
{
cout << "Not enough arguments! Stop run. " << endl;
return -1;
}
/*
we're going to make a histogram, and set some parameters about it's X and Y axes
*/
TH1D *nuflavorint_hist = new TH1D("nuflavorint", "",3,1,4);
nuflavorint_hist->SetTitle("Neutrino Flavors");
nuflavorint_hist->GetXaxis()->SetTitle("Neutrino Flavors (1=e, 2=muon, 3=tau)");
nuflavorint_hist->GetYaxis()->SetTitle("Weigthed Fraction of Total Detected Events");
nuflavorint_hist->GetXaxis()->SetTitleOffset(1.2);
nuflavorint_hist->GetYaxis()->SetTitleOffset(1.2);
nuflavorint_hist->GetXaxis()->CenterTitle();
nuflavorint_hist->GetYaxis()->CenterTitle();
for(int i=1; i < argc; i++)
{ // loop over the input files
//now we are going to load the icefinal.root file and draw in the "passing_events" tree, which stores info
string readfile = string(argv[i]);
TFile *AnitaFile = new TFile(( readfile ).c_str());
cout << "AnitaFile" << endl;
TTree *passing_events = (TTree*)AnitaFile->Get("passing_events");
cout << "Reading AnitaFile..." << endl;
//declare three variables we are going to use later
int num_pass; // number of entries (ultra-neutrinos);
double weight; // weight of neutrino counts;
int nuflavorint; // neutrino flavors;
num_pass = passing_events->GetEntries();
cout << "num_pass is " << num_pass << endl;
/*PRIMARIES VARIABLES*/
//set the "branch" of the tree which stores specific pieces of information
passing_events->SetBranchAddress("weight", &weight);
passing_events->SetBranchAddress("nuflavor", &nuflavorint);
//loop over all the events in the tree
for (int k=0; k <=num_pass; k++)
{
passing_events->GetEvent(k);
nuflavorint_hist->Fill(nuflavorint, weight); //fill the histogram with this value and this weight
} // CLOSE FOR LOOP OVER NUMBER OF EVENTS
} // CLOSE FOR LOOP OVER NUMBER OF INPUT FILES
//set up some parameters to make things loo pretty
gStyle->SetHistFillColor(0);
gStyle->SetHistFillStyle(1);
gStyle->SetHistLineColor(1);
gStyle->SetHistLineStyle(0);
gStyle->SetHistLineWidth(2.5); //Setup plot Style
//make a "canvas" to draw on
TCanvas *c4 = new TCanvas("c4", "nuflavorint", 1100,850);
gStyle->SetOptTitle(1);
gStyle->SetStatX(0.33);
gStyle->SetStatY(0.87);
nuflavorint_hist->Draw("HIST"); //draw on it
//Save Plots
//make the line thicker and then save the result
gStyle->SetHistLineWidth(9);
c4->SaveAs("nuflavorint.png");
gStyle->SetHistLineWidth(2);
c4->SaveAs("nuflavorint.pdf");
delete c4; //clean up
return 0; //return successfully
}
|
| Attachment 4: bashrc_anita.sh
|
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
module load cmake/3.11.4
module load gnu/7.3.0
export CC=`which gcc`
export CXX=`which g++`
export BOOST_ROOT=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/boost_build
export LD_LIBRARY_PATH=${BOOST_ROOT}/stage/lib:$LD_LIBRARY_PATH
export BOOST_LIB=$BOOST_ROOT/stage/lib
export LD_LIBRARY_PATH=$BOOST_LIB:$LD_LIBRARY_PATH
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
eval 'source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh'
|
| Attachment 5: getting_running_with_anita_stuff.pdf
|
| Attachment 6: getting_running_with_anita_stuff.pptx
|
| Attachment 7: test_plot.mk
|
# Makefile for the ROOT test programs. # This Makefile shows nicely how to compile and link applications
# using the ROOT libraries on all supported platforms.
#
# Copyright (c) 2000 Rene Brun and Fons Rademakers
#
# Author: Fons Rademakers, 29/2/2000
include Makefile.arch
################################################################################
# Site specific flags
################################################################################
# Toggle these as needed to get things to install
#BOOSTFLAGS = -I boost_1_48_0
# commented out for kingbee and older versions of gcc
ANITA3_EVENTREADER=1
# Uncomment to enable healpix
#USE_HEALPIX=1
# Uncomment to disable explicit vectorization (but will do nothing if ANITA_UTIL is not available)
#VECTORIZE=1
# The ROOT flags are added to the CXXFLAGS in the .arch file
# so this should be simpler...
ifeq (,$(findstring -std=, $(CXXFLAGS)))
ifeq ($(shell test $(GCC_MAJOR) -lt 5; echo $$?),0)
ifeq ($(shell test $(GCC_MINOR) -lt 5; echo $$?),0)
CXXFLAGS += -std=c++0x
else
CXXFLAGS += -std=c++11
endif
endif
endif
################################################################################
# If not compiling with C++11 (or later) support, all occurrences of "constexpr"
# must be replaced with "const", because "constexpr" is a keyword
# which pre-C++11 compilers do not support.
# ("constexpr" is needed in the code to perform in-class initialization
# of static non-integral member objects, i.e.:
# static const double c_light = 2.99e8;
# which works in C++03 compilers, must be modified to:
# static constexpr double c_light = 2.99e8;
# to work in C++11, but adding "constexpr" breaks C++03 compatibility.
# The following compiler flag defines a preprocessor macro which is
# simply:
# #define constexpr const
# which replaces all instances of the text "constexpr" and replaces it
# with "const".
# This preserves functionality while only affecting very specific semantics.
ifeq (,$(findstring -std=c++1, $(CXXFLAGS)))
CPPSTD_FLAGS = -Dconstexpr=const
endif
# Uses the standard ANITA environment variable to figure
# out if ANITA libs are installed
ifdef ANITA_UTIL_INSTALL_DIR
ANITA_UTIL_EXISTS=1
ANITA_UTIL_LIB_DIR=${ANITA_UTIL_INSTALL_DIR}/lib
ANITA_UTIL_INC_DIR=${ANITA_UTIL_INSTALL_DIR}/include
LD_ANITA_UTIL=-L$(ANITA_UTIL_LIB_DIR)
LIBS_ANITA_UTIL=-lAnitaEvent -lRootFftwWrapper
INC_ANITA_UTIL=-I$(ANITA_UTIL_INC_DIR)
ANITA_UTIL_ETC_DIR=$(ANITA_UTIL_INSTALL_DIR)/etc
endif
ifdef ANITA_UTIL_EXISTS
CXXFLAGS += -DANITA_UTIL_EXISTS
endif
ifdef VECTORIZE
CXXFLAGS += -DVECTORIZE -march=native -fabi-version=0
endif
ifdef ANITA3_EVENTREADER
CXXFLAGS += -DANITA3_EVENTREADER
endif
ifdef USE_HEALPIX
CXXFLAGS += -DUSE_HEALPIX `pkg-config --cflags healpix_cxx`
LIBS += `pkg-config --libs healpix_cxx`
endif
################################################################################
GENERAL_FLAGS = -g -O2 -pipe -m64 -pthread
WARN_FLAGS = -W -Wall -Wextra -Woverloaded-virtual
# -Wno-unused-variable -Wno-unused-parameter -Wno-unused-but-set-variable
CXXFLAGS += $(GENERAL_FLAGS) $(CPPSTD_FLAGS) $(WARN_FLAGS) $(ROOTCFLAGS) $(INC_ANITA_UTIL)
DBGFLAGS = -pipe -Wall -W -Woverloaded-virtual -g -ggdb -O0 -fno-inline
DBGCXXFLAGS = $(DBGFLAGS) $(ROOTCFLAGS) $(BOOSTFLAGS)
LDFLAGS += $(CPPSTD_FLAGS) $(LD_ANITA_UTIL) -I$(BOOST_ROOT) -L.
LIBS += $(LIBS_ANITA_UTIL)
# Mathmore not included in the standard ROOT libs
LIBS += -lMathMore
DICT = classdict
OBJS = vector.o position.o earthmodel.o balloon.o icemodel.o signal.o ray.o Spectra.o anita.o roughness.o secondaries.o Primaries.o Tools.o counting.o $(DICT).o Settings.o Taumodel.o screen.o GlobalTrigger.o ChanTrigger.o SimulatedSignal.o EnvironmentVariable.o source.o random.o
BINARIES = test_plot$(ExeSuf)
################################################################################
.SUFFIXES: .$(SrcSuf) .$(ObjSuf) .$(DllSuf)
all: $(BINARIES)
$(BINARIES): %: %.$(SrcSuf) $(OBJS)
$(LD) $(CXXFLAGS) $(LDFLAGS) $(OBJS) $< $(LIBS) $(OutPutOpt) $@
@echo "$@ done"
.PHONY: clean
clean:
@rm -f $(BINARIES)
%.$(ObjSuf) : %.$(SrcSuf) %.h
@echo "<**Compiling**> "$<
$(LD) $(CXXFLAGS) -c $< -o $@
|
|
38
|
Wed May 15 00:38:54 2019 |
Brian Clark | Get a quick start with AraSim on osc | Software | Follow the instructions in the attached "getting_started_with_ara.pdf" file to download AraSim, compile it, generate results, and plot those results. |
| Attachment 1: plotting_example.cc
|
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include <cstdio>
#include <cstdlib>
#include <unistd.h>
#include <cstring>
#include <unistd.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TH1.h"
#include "TF1.h"
#include "TF2.h"
#include "TFile.h"
#include "TRandom.h"
#include "TRandom2.h"
#include "TRandom3.h"
#include "TTree.h"
#include "TLegend.h"
#include "TLine.h"
#include "TROOT.h"
#include "TPostScript.h"
#include "TCanvas.h"
#include "TH2F.h"
#include "TText.h"
#include "TProfile.h"
#include "TGraphErrors.h"
#include "TStyle.h"
#include "TMath.h"
#include "TVector3.h"
#include "TRotation.h"
#include "TSpline.h"
#include "Math/InterpolationTypes.h"
#include "Math/Interpolator.h"
#include "Math/Integrator.h"
#include "TGaxis.h"
#include "TPaveStats.h"
#include "TLatex.h"
#include "Constants.h"
#include "Settings.h"
#include "Position.h"
#include "EarthModel.h"
#include "Tools.h"
#include "Vector.h"
#include "IceModel.h"
#include "Trigger.h"
#include "Spectra.h"
#include "signal.hh"
#include "secondaries.hh"
#include "Ray.h"
#include "counting.hh"
#include "Primaries.h"
#include "Efficiencies.h"
#include "Event.h"
#include "Detector.h"
#include "Report.h"
using namespace std;
/////////////////Plotting Script for nuflavorint for AraSIMQC
/////////////////Created by Kaeli Hughes, modified by Brian Clark
/////////////////Prepared on April 28 2016 as an "introductor script" to plotting with AraSim; for new users of queenbee
class EarthModel;
int main(int argc, char **argv)
{
gStyle->SetOptStat(111111); //this is a selection of statistics settings; you should do some googling and figure out exactly what this particular comibation does
gStyle->SetOptDate(1); //this tells root to put a date and timestamp on whatever plot we output
if(argc<2){ //if you don't have at least 1 file to run over, then you haven't given it a file to analyze; this checks this
cout<<"Not enough arguments! Abort run."<<endl;
}
//Create the histogram
TCanvas *c2 = new TCanvas("c2", "nuflavorint", 1100,850); //make a canvas on which to plot the data
TH1F *nuflavorint_hist = new TH1F("nuflavorint_hist", "nuflavorint histogram", 3, 0.5, 3.5); //create a histogram with three bins
nuflavorint_hist->GetXaxis()->SetNdivisions(3); //set the number of divisions
int total_thrown=0; // a variable to hold the grant total number of events thrown for all input files
for(int i=1; i<argc;i++){ //loop over the input files
string readfile; //create a variable called readfile that will hold the title of the simulation file
readfile = string(argv[i]); //set the readfile variable equal to the filename
Event *event = 0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
Report *report=0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
Settings *settings = 0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
TFile *AraFile=new TFile(( readfile ).c_str()); //make a new file called "AraFile" that will be simulation file we are reading in
if(!(AraFile->IsOpen())) return 0; //checking to see if the file we're trying to read in opened correctly; if not, bounce out of the program
int num_pass;//number of passing events
TTree *AraTree=(TTree*)AraFile->Get("AraTree"); //get the AraTree
TTree *AraTree2=(TTree*)AraFile->Get("AraTree2"); //get the AraTree2
AraTree2->SetBranchAddress("event",&event); //get the event branch
AraTree2->SetBranchAddress("report",&report); //get the report branch
AraTree->SetBranchAddress("settings",&settings); //get the settings branch
num_pass=AraTree2->GetEntries(); //get the number of passed events in the data file
AraTree->GetEvent(0); //get the first event; sometimes the tree does not instantiate properly if you'd explicitly "active" the first event
total_thrown+=(settings->NNU); //add the number of events from this file (The NNU variable) to the grand total; NNU is the number of THROWN neutrinos
for (int k=0; k<num_pass; k++){ //going to fill the histograms for as many events as were in this input file
AraTree2->GetEvent(k); //get the even from the tree
int nuflavorint; //make the container variable
double weight; //the weight of the event
int trigger; //the global trigger value for the event
nuflavorint=event->nuflavorint; //draw the event out; one of the objects in the event class is the nuflavorint, and this is the syntax for accessing it
weight=event->Nu_Interaction[0].weight; //draw out the weight of the event
trigger=report->stations[0].Global_Pass; //find out if the event was a triggered event or not
/*
if(trigger!=0){ //check if the event triggered
nuflavorint_hist->Fill(nuflavorint,weight); //fill the event into the histogram; the first argument of the fill (which is mandatory) is the value you're putting into the histogram; the second value is optional, and is the weight of the event in the histogram
//in this particular version of the code then, we are only plotting the TRIGGERED events; if you wanted to plot all of the events, you could instead remove this "if" condition and just Fill everything
}
*/
nuflavorint_hist->Fill(nuflavorint,weight); //fill the event into the histogram; the first argument of the fill (which is mandatory)
}
}
//After looping over all the files, make the plots and save them
//do some stuff to get the "total events thrown" text box ready; this is useful because on the plot itself you can then see how many events you THREW into the simulation; this is especially useful if you're only plotting passed events, but want to know what fraction of your total thrown that is
char *buffer= (char*) malloc (250); //declare a buffer pointer, and allocate it some chunk of memory
int a = snprintf(buffer, 250,"Total Events Thrown: %d",total_thrown); //print the words "Total Events Thrown: %d" to the variable "buffer" and tell the system how long that phrase is; the %d sign tells C++ to replace that "%d" with the next argument, or in this case, the number "total_thrown"
if(a>=250){ //if the phrase is longer than the pre-allocated space, increase the size of the buffer until it's big enough
buffer=(char*) realloc(buffer, a+1);
snprintf(buffer, 250,"Total Events Thrown: %d",total_thrown);
}
TLatex *u = new TLatex(.3,.01,buffer); //create a latex tex object which we can draw on the cavas
u->SetNDC(kTRUE); //changes the coordinate system for the tex object plotting
u->SetIndiceSize(.1); //set the size of the latex index
u->SetTextSize(.025); //set the size of the latex text
nuflavorint_hist->Draw(); //draw the histogram
nuflavorint_hist->GetXaxis()->SetTitle("Neutrino Flavor"); //set the x-axis label
nuflavorint_hist->GetYaxis()->SetTitle("Number of Events (weighted)"); //set the y-axis label
nuflavorint_hist->GetYaxis()->SetTitleOffset(1.5); //set the separation between the y-axis and it's label; root natively makes this smaller than is ideal
nuflavorint_hist->SetLineColor(kBlack); //set the color of the histogram to black, instead of root's default navy blue
u->Draw(); //draw the statistics box information
c2->SaveAs("outputs/plotting_example.png"); //save the canvas as a JPG file for viewing
c2->SaveAs("outputs/plotting_example.pdf"); //save the canvas as a PDF file for viewing
c2->SaveAs("outputs/plotting_example.root"); //save the canvas as a ROOT file for viewing or editing later
} //end main; this is the end of the script
|
| Attachment 2: plotting_example.mk
|
#############################################################################
## Makefile -- New Version of my Makefile that works on both linux
## and mac os x
## Ryan Nichol <rjn@hep.ucl.ac.uk>
##############################################################################
##############################################################################
##############################################################################
##
##This file was copied from M.readGeom and altered for my use 14 May 2014
##Khalida Hendricks.
##
##Modified by Brian Clark for use on plotting_example on 28 April 2016
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##
##############################################################################
##############################################################################
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o plotting_example.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc plotting_example.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = plotting_example
all : $(PROGRAMS)
plotting_example : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
| Attachment 3: test_setup.txt
|
NFOUR=1024
EXPONENT=21
NNU=300 // number of neutrino events
NNU_PASSED=10 // number of neutrino events that are allowed to pass the trigger
ONLY_PASSED_EVENTS=0 // 0 (default): AraSim throws NNU events whether or not they pass; 1: AraSim throws events until the number of events that pass the trigger is equal to NNU_PASSED (WARNING: may cause long run times if reasonable values are not chosen)
NOISE_WAVEFORM_GENERATE_MODE=0 // generate new noise waveforms for each events
NOISE_EVENTS=16 // number of pure noise waveforms
TRIG_ANALYSIS_MODE=0 // 0 = signal + noise, 1 = signal only, 2 = noise only
DETECTOR=1 // ARA stations 1 to 7
NOFZ=1
core_x=10000
core_y=10000
TIMESTEP=5.E-10 // value for 2GHz actual station value
TRIG_WINDOW=1.E-7 // 100ns which is actual testbed trig window
POWERTHRESHOLD=-6.06 // 100Hz global trig rate for 3 out of 16 ARA stations
POSNU_RADIUS=3000
V_MIMIC_MODE=0 // 0 : global trig is located center of readout windows
DATA_SAVE_MODE=0 // 2 : don't save any waveform informations at all
DATA_LIKE_OUTPUT=0 // 0 : don't save any waveform information to eventTree
BORE_HOLE_ANTENNA_LAYOUT=0
SECONDARIES=0
TRIG_ONLY_BH_ON=0
CALPULSER_ON=0
USE_MANUAL_GAINOFFSET=0
USE_TESTBED_RFCM_ON=0
NOISE_TEMP_MODE=0
TRIG_THRES_MODE=0
READGEOM=0 // reads geometry information from the sqlite file or not (0 : don't read)
TRIG_MODE=0 // use vpol, hpol separated trigger mode. by default N_TRIG_V=3, N_TRIG_H=3. You can change this values
number_of_stations=1
core_x=10000
core_y=10000
DETECTOR=1
DETECTOR_STATION=2
DATA_LIKE_OUTPUT=0
NOISE_WAVEFORM_GENERATE_MODE=0 // generates new waveforms for every event
NOISE=0 //flat thermal noise
NOISE_CHANNEL_MODE=0 //using different noise temperature for each channel
NOISE_EVENTS=16 // number of noise events which will be store in the trigger class for later use
ANTENNA_MODE=1
APPLY_NOISE_FIGURE=0
|
| Attachment 4: bashrc_anita.sh
|
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
module load cmake/3.11.4
module load gnu/7.3.0
export CC=`which gcc`
export CXX=`which g++`
export BOOST_ROOT=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/boost_build
export LD_LIBRARY_PATH=${BOOST_ROOT}/stage/lib:$LD_LIBRARY_PATH
export BOOST_LIB=$BOOST_ROOT/stage/lib
export LD_LIBRARY_PATH=$BOOST_LIB:$LD_LIBRARY_PATH
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
eval 'source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh'
|
| Attachment 5: sample_bash_profile.sh
|
# .bash_profile
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
# User specific environment and startup programs
PATH=$PATH:$HOME/.local/bin:$HOME/bin
export PATH
|
| Attachment 6: sample_bashrc.sh
|
# .bashrc
source bashrc_anita.sh
# we also want to set two more environment variables that ANITA needs
# you should update ICEMC_SRC_DIR and ICEMC_BUILD_DIR to wherever you
# downloaded icemc too
export ICEMC_SRC_DIR=/path/to/icemc #change this line!
export ICEMC_BUILD_DIR=/path/to/icemc #change this line!
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
|
| Attachment 7: getting_running_with_ara_stuff.pdf
|
| Attachment 8: getting_running_with_ara_stuff.pptx
|
|
26
|
Sun Aug 26 19:23:57 2018 |
Brian Clark | Get a quick start with AraSim on OSC Oakley | Software | These are instructions I wrote for Rishabh Khandelwal to facilitate a "fast" start on Oakley at OSC. It was to help him run AraSim in batch jobs on Oakley.
It basically has you use software dependencies that I pre-installed on my OSC account at /users/PAS0654/osu0673/PhasedArraySimulation.
It also gives a "batch_processing" folder with examples for how to successfully run AraSim batch jobs (with correct output file management) on Oakley.
Sourcing these exact dependencies will not work on Owens or Ruby, sorry. |
| Attachment 1: forRishabh.tar.gz
|
|
20
|
Tue Mar 20 09:24:37 2018 |
Brian Clark | Get Started with Making Plots for IceMC | Software | |
Second, here is the page for the software IceMC, which is the Monte Carlo software for simulating neutrinos for ANITA.
On that page are good instructions for downloading the software and how to run it. You will have the choice of running it on a (1) a personal machine (if you want to use your personal mac or linux machine), (2) a queenbee laptop in the lab, or (3) on a kingbee account which I will send an email about shortly. Running IceMC will require a piece of statistics software called ROOT that can be somewhat challenging to install--it is already installed on Kingbee and OSC, so it is easier to get started there. If you want to use Kingbee, just try downloading and running. If you want to use OSC, you're first going to need to follow instructions to access a version installed on OSC. Still getting that together.
So, familiarize yourself with the command line, and then see if you can get ROOT and IceMC installed and running. Then plots. |
|
25
|
Wed Aug 1 11:37:10 2018 |
Andres Medina | Flux Ordering | Hardware | Bought 951 Non-Resin Soldering Flux. This is the preferred variety. This could be found on this website https://www.kester.com/products/product/951-soldering-flux.
The amount of Flux that was bought was 1 Gallon (Lasts quite some time). The price was $83.86 with approximate shipping of $43. This was done with a Pcard and a tax exempt form.
The website used to purchase this was https://www.alliedelec.com/kester-solder-63-0000-0951/70177935/ |
|
17
|
Mon Nov 20 08:31:48 2017 |
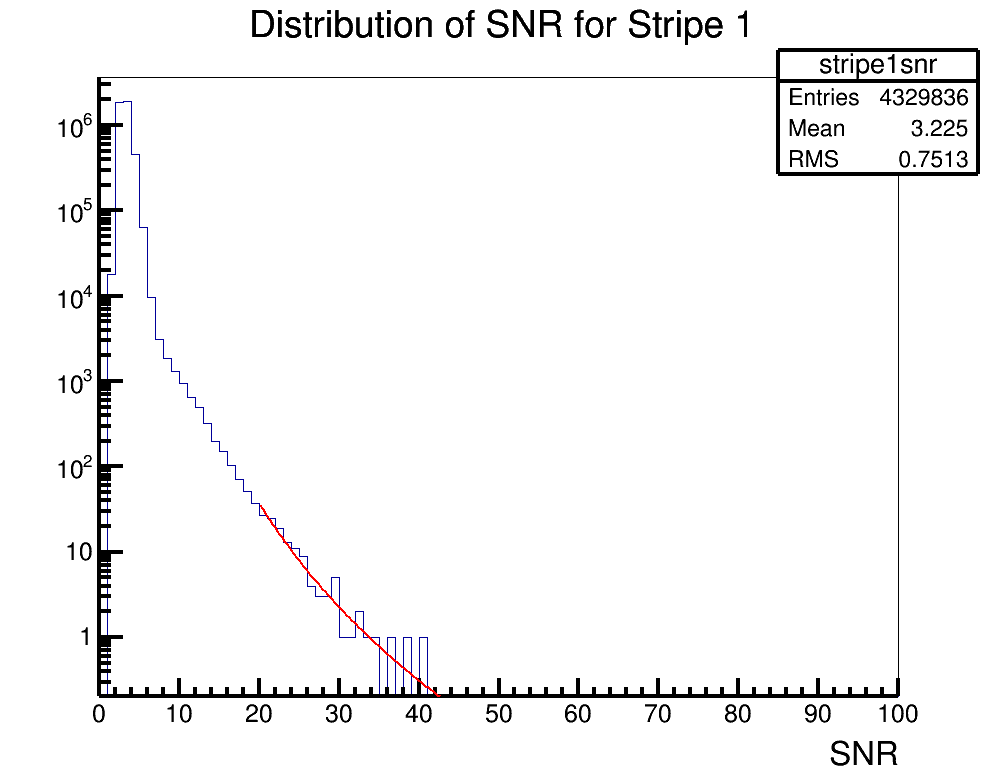
Brian Clark and Oindree Banerjee | Fit a Function in ROOT | Analysis | Sometimes you need to fit a function to a histogram in ROOT. Attached is code for how to do that in the simple case of a power law fit.
To run the example, you should type "root fitSnr.C" in the command line. The code will access the source histogram (hstrip1nsr.root, which is actually ANITA-2 satellite data). The result is stripe1snrfit.png. |
| Attachment 1: fitSnr.C
|
#include "TF1.h"
void fitSnr();
void fitSnr()
{
gStyle->SetLineWidth(4); //set some style parameters
TFile *stripe1file = new TFile("hstripe1snr.root"); //import the file containing the histogram to be fit
TH1D *hstripe1 = (TH1D*)stripe1file->Get("stripe1snr"); //get the histogram to be fit
TCanvas c1("c1","c1",1000,800); //make a canvas
hstripe1->Draw(""); //draw it
c1.SetLogy(); //set a log axis
//need to declare an equation
//I want to fit for two paramters, in the equation these are [0] and [1]
//so, you'll need to re-write the equation to whatever you're trying to fit for
//but ROOT wants the variables to find to be given as [0], [1], [2], etc.
char equation1[150]; //declare a container for the equation
sprintf(equation1,"([0]*(x^[1]))"); //declare the equation
TF1 *fit1 = new TF1("PowerFit",equation1,20,50); //create a function to fit with, with the range being 20 to 50
//now, we need to set the initial parameters of the fit
//fit->SetParameter(0,H->GetRMS()); //this should be a good starting place for a standard deviation like variable
//fit->SetParameter(1,H->GetMaximum()); //this should be a good starting place for amplitude like variable
fit1->SetParameter(0,60000.); //for our example, we will manually choose this
fit1->SetParameter(1,-3.);
hstripe1->Fit("PowerFit","R"); //actually do the fit;
fit1->Draw("same"); //draw the fit
//now, we want to print out some parameters to see how good the fit was
cout << "par0 " << fit1->GetParameter(0) << " par1 " << fit1->GetParameter(1) << endl;
cout<<"chisquare "<<fit1->GetChisquare()<<endl;
cout<<"Ndf "<<fit1->GetNDF()<<endl;
cout<<"reduced chisquare "<<double(fit1->GetChisquare())/double(fit1->GetNDF())<<endl;
cout<<" "<<endl;
c1.SaveAs("stripe1snrfit.png");
}
|
| Attachment 2: hstripe1snr.root
|
| Attachment 3: stripe1snrfit.png
|  |
|
16
|
Thu Oct 26 08:44:58 2017 |
Brian Clark | Find Other Users on OSC | Other | Okay, so our group has two "project" spaces on OSC (the Ohio Supercomputer). The first is for Amy's group, and is a project workspace called "PAS0654". The second is the CCAPP Condo (literally, CCAPP has some pre-specified rental time, and hence "condo") on OSC, and this is project PCON0003.
When you are signed up for the supercomputer, one of two things happen:
- You will be given a username under the PAS0654 group, and in which case, your username will be something like osu****. Connolly home drive is /users/PAS0654/osu****. Beatty home drive is /users/PAS0174/osu****. CCAPP home drive is /users/PCON0003/pcon****.
- You will be given a username under the PCON0003 group, and in which case, your username will be something like cond****.
If you are given a osu**** username, you must make sure to be added to the CCAPP Condo so that you can use Ruby compute resources. It will not be automatic.
Some current group members, and their OSC usernames. In parenthesis is the project space they are found in.
Current Users
osu0673: Brian Clark (PAS0654)
cond0068: Jorge Torres-Espinosa (PCON0003)
osu8619: Keith McBride (PAS0174)
osu9348: Julie Rolla (PAS0654)
osu9979: Lauren Ennesser (PAS0654)
osu6665: Amy Connolly (PAS0654)
Past Users
osu0426: Oindree Banerjee (PAS0654)
osu0668: Brian Dailey (PAS0654)
osu8620: Jacob Gordon (PAS0174)
osu8386: Sam Stafford (ANITA analysis in /fs/scratch/osu8386) (PAS0174)
cond0091: Judge Rajasekera (PCON0003) |
|
1
|
Thu Mar 16 09:01:50 2017 |
Amy Connolly | Elog instructions | Other | Log into kingbee.mps.ohio-state.edu first, then log into radiorm.physics.ohio-state.edu.
From Keith Stewart 03/16/17: It appears that radiorm SSH from offsite is closed. So you will need to be on an OSU network physically or via VPN. fox is also blocked from offsite as well. Kingbee should still be available for now. If you want to use it as a jump host to get to radiorm without VPN. However, you will want to get comfortable with the VPN before it is a requirement.
Carl 03/16/17: I could log in even while using a hard line and plugged in directly to the network.
From Bryan Dunlap 12/16/17: I have set up the group permissions on the elog directory so you and your other designated people can edit files. I have configured sudo to allow you all to restart the elogd service. Once you have edited the file [/home/elog/elog.cfg I think], you can then type
sudo /sbin/service elogd restart
to restart the daemon so it re-reads the config. Sudo will prompt you for your password before it executes the command. |
|
48
|
Thu Jun 8 16:29:45 2023 |
Alan Salcedo | Doing IceCube/ARA coincidence analysis | | These documents contain information on how to run IceCube/ARA coincidence simulations and analysis. All technical information of where codes are stored and how to use them is detailed in the technical note. Other supportive information for physics understanding is in the powerpoint slides. The technical note will direct you to other documents in this elog in the places where you may need supplemental information. |
| Attachment 1: IceCube_ARA_Coincidence_Analysis___Technical_Note.pdf
|
| Attachment 2: ICARA_Coincident_Events_Introduction.pptx
|
| Attachment 3: ICARA_Analysis_Template.ipynb
|
{
"cells": [
{
"cell_type": "markdown",
"id": "bcdfb138",
"metadata": {},
"source": [
"# IC/ARA Coincident Simulation Events Analysis"
]
},
{
"cell_type": "markdown",
"id": "c7ad7c80",
"metadata": {},
"source": [
"### Settings and imports"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "b6915a86",
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"<style>.container { width:75% !important; }</style>"
],
"text/plain": [
"<IPython.core.display.HTML object>"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"## Makes this notebook maximally wide\n",
"from IPython.display import display, HTML\n",
"display(HTML(\"<style>.container { width:75% !important; }</style>\"))"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "6ef9e20b",
"metadata": {},
"outputs": [],
"source": [
"## Author: Alex Machtay (machtay.1@osu.edu)\n",
"## Modified by: Alan Salcedo (salcedogomez.1@osu.edu)\n",
"## Date: 4/26/23\n",
"\n",
"## Purpose:\n",
"### This script will read the data files produced by AraRun_corrected_MultStat.py to make histograms relevant plots of\n",
"### neutrino events passing through icecube and detected by ARA station (in AraSim)\n",
"\n",
"\n",
"## Imports\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"import sys\n",
"sys.path.append(\"/users/PAS0654/osu8354/root6_18_build/lib\") # go to parent dir\n",
"sys.path.append(\"/users/PCON0003/cond0068/.local/lib/python3.6/site-packages\")\n",
"import math\n",
"import argparse\n",
"import glob\n",
"import pandas as pd\n",
"pd.options.mode.chained_assignment = None # default='warn'\n",
"from mpl_toolkits.mplot3d import Axes3D\n",
"import jupyterthemes as jt"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "f24b8292",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"/bin/bash: jt: command not found\r\n"
]
}
],
"source": [
"## Set style for the jupyter notebook\n",
"!jt -t grade3 -T -N -kl -lineh 160 -f code -fs 14 -ofs 14 -cursc o\n",
"jt.jtplot.style('grade3', gridlines='')"
]
},
{
"cell_type": "markdown",
"id": "a5a812d6",
"metadata": {},
"source": [
"### Set constants\n",
"#### These are things like the position of ARA station holes, the South Pole, IceCube's position in ARA station's coordinates, and IceCube's radius"
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "cdb851de",
"metadata": {},
"outputs": [],
"source": [
"## What IceCube (IC) station are you analyzing\n",
"station = 1\n",
"\n",
"## What's the radius around ARA where neutrinos were injected\n",
"inj_rad = 5 #in km\n",
"\n",
"## IceCube's center relative to each ARA station\n",
"IceCube = [[-1128.08, -2089.42, -1942.39], [-335.812, -3929.26, -1938.23],\n",
" [-2320.67, -3695.78, -1937.35], [-3153.04, -1856.05, -1942.81], [472.49, -5732.98, -1922.06]] #IceCube's position relative to A1, A2, or A3\n",
"\n",
"#To calculate this, we need to do some coordinate transformations. Refer to this notebook to see the calculations: \n",
"# IceCube_Relative_to_ARA_Stations.ipynb (found here - http://radiorm.physics.ohio-state.edu/elog/How-To/48) \n",
"\n",
"## IceCube's radius\n",
"IceCube_radius = 564.189583548 #Modelling IceCube as a cylinder, we find the radius with V = h * pi*r^2 with V = 1x10^9 m^3 and h = 1x10^3 m "
]
},
{
"cell_type": "markdown",
"id": "af5c6fc2",
"metadata": {},
"source": [
"### Read the data\n",
"\n",
"#### Once we import the data, we'll make dataframes to concatenate it and make some calculations"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "921f61f8",
"metadata": {},
"outputs": [],
"source": [
"## Import data files\n",
"\n",
"#Here, it's from OSC Connolly's group project space\n",
"source = '/fs/project/PAS0654/IceCube_ARA_Coincident_Search/AraSim/outputs/Coincident_Search_Runs/20M_GZK_5km_S1_correct' \n",
"num_files = 200 # Number of files to read in from the source directory\n",
"\n",
"## Make a list of all of the paths to check \n",
"file_list = []\n",
"for i in range(1, num_files + 1):\n",
" for name in glob.glob(source + \"/\" + str(i) + \"/*.csv\"):\n",
" file_list.append(str(name))\n",
" #file_list gets paths to .csv files\n",
" \n",
"## Now read the csv files into a pandas dataframe\n",
"dfs = []\n",
"for filename in file_list:\n",
" df = pd.read_csv(filename, index_col=None, header=0) #Store each csv file into a pandas data frame\n",
" dfs.append(df) #Append the csv file to store all of them in one\n",
"frame = pd.concat(dfs, axis=0, ignore_index = True) #Concatenate pandas dataframes "
]
},
{
"cell_type": "markdown",
"id": "813fb3ee",
"metadata": {},
"source": [
"### Work with the data\n",
"\n",
"#### All the data from our coincidence simulations (made by AraRun_MultStat.sh) is now stored in a pandas data frame that we can work with"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "8b449583",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"19800000\n",
"19799802\n"
]
}
],
"source": [
"## Now let's clean up our data and calculate other relevant things\n",
"print(len(frame))\n",
"frame = frame[frame['weight'].between(0,1)] #Filter out events with ill-defined weight (should be between 0 and 1)\n",
"print(len(frame))\n",
"\n",
"frame['x-positions'] = (frame['Radius (m)'] * np.cos(frame['Azimuth (rad)']) * np.sin(frame['Zenith (rad)']))\n",
"frame['y-positions'] = (frame['Radius (m)'] * np.sin(frame['Azimuth (rad)']) * np.sin(frame['Zenith (rad)']))\n",
"frame['z-positions'] = (frame['Radius (m)'] * np.cos(frame['Zenith (rad)']))\n",
"\n",
"## The energy in eV will be 10 raised to the number in the file, multiplied by 1-y (y is inelasticity)\n",
"frame['Nu Energies (eV)'] = np.power(10, (frame['Energy (log10) (eV)']))\n",
"frame['Mu Energies (eV)'] = ((1-frame['Inelasticity']) * frame['Nu Energies (eV)']) #Energy of the produced lepton\n",
"#Here the lepton is not a muon necesarily, hence the label 'Mu Energies (eV)' may be misleading\n",
"\n",
"## Get a frame with only coincident events\n",
"coincident_frame = frame[frame['Coincident'] == 1] \n",
"\n",
"## And a frame for strictly events *detected* by ARA\n",
"detected_frame = frame[frame['Detected'] == 1]\n",
"\n",
"\n",
"## Now let's calculate the energy of the lepton when reaching IceCube (IC)\n",
"\n",
"# To do this correctly, I need to find exactly the distance traveled by the muon and apply the equation\n",
"# I need the trajectory of the muon to find the time it takes to reach IceCube, then I can find the distance it travels in that time\n",
"# I should allow events that occur inside the icecube volume to have their full energy (but pretty much will happen anyway)\n",
"## a = sin(Theta)*cos(Phi)\n",
"## b = sin(Theta)*sin(Phi)\n",
"## c = cos(Theta)\n",
"## a_0 = x-position\n",
"## b_0 = y-position\n",
"## c_0 = z-position\n",
"## x_0 = IceCube[0]\n",
"## y_0 = IceCube[1]\n",
"## z_0 = IceCube[2]\n",
"## t = (-(a*(a_0-x_0) + b*(b_0-y_0))+D**0.5)/(a**2+b**2)\n",
"## D = (a**2+b**2)*R_IC**2 - (a*(b_0-y_0)+b*(a_0-x_0))**2\n",
"## d = ((a*t)**2 + (b*t)**2 + (c*t)**2)**0.5\n",
"\n",
"## Trajectories\n",
"coincident_frame['a'] = (np.sin(coincident_frame['Theta (rad)'])*np.cos(coincident_frame['Phi (rad)']))\n",
"coincident_frame['b'] = (np.sin(coincident_frame['Theta (rad)'])*np.sin(coincident_frame['Phi (rad)']))\n",
"coincident_frame['c'] = (np.cos(coincident_frame['Theta (rad)']))\n",
"\n",
"## Discriminant\n",
"coincident_frame['D'] = ((coincident_frame['a']**2 + coincident_frame['b']**2)*IceCube_radius**2 - \n",
" (coincident_frame['a']*(coincident_frame['y-position (m)']-IceCube[station-1][1])- ## I think this might need to be a minus sign!\n",
" coincident_frame['b']*(coincident_frame['x-position (m)']-IceCube[station-1][0]))**2)\n",
"\n",
"## Interaction time (this is actually the same as the distance traveled, at least for a straight line)\n",
"coincident_frame['t_1'] = (-(coincident_frame['a']*(coincident_frame['x-position (m)']-IceCube[station-1][0])+\n",
" coincident_frame['b']*(coincident_frame['y-position (m)']-IceCube[station-1][1]))+\n",
" np.sqrt(coincident_frame['D']))/(coincident_frame['a']**2+coincident_frame['b']**2)\n",
"coincident_frame['t_2'] = (-(coincident_frame['a']*(coincident_frame['x-position (m)']-IceCube[station-1][0])+\n",
" coincident_frame['b']*(coincident_frame['y-position (m)']-IceCube[station-1][1]))-\n",
" np.sqrt(coincident_frame['D']))/(coincident_frame['a']**2+coincident_frame['b']**2)\n",
"\n",
"## Intersection coordinates\n",
"coincident_frame['x-intersect_1'] = (coincident_frame['a'] * coincident_frame['t_1'] + coincident_frame['x-position (m)'])\n",
"coincident_frame['y-intersect_1'] = (coincident_frame['b'] * coincident_frame['t_1'] + coincident_frame['y-position (m)'])\n",
"coincident_frame['z-intersect_1'] = (coincident_frame['c'] * coincident_frame['t_1'] + coincident_frame['z-position (m)'])\n",
"\n",
"coincident_frame['x-intersect_2'] = (coincident_frame['a'] * coincident_frame['t_2'] + coincident_frame['x-position (m)'])\n",
"coincident_frame['y-intersect_2'] = (coincident_frame['b'] * coincident_frame['t_2'] + coincident_frame['y-position (m)'])\n",
"coincident_frame['z-intersect_2'] = (coincident_frame['c'] * coincident_frame['t_2'] + coincident_frame['z-position (m)'])\n",
"\n",
"## Distance traveled (same as the parametric time, at least for a straight line)\n",
"coincident_frame['d_1'] = (np.sqrt((coincident_frame['a']*coincident_frame['t_1'])**2+\n",
" (coincident_frame['b']*coincident_frame['t_1'])**2+\n",
" (coincident_frame['c']*coincident_frame['t_1'])**2))\n",
"coincident_frame['d_2'] = (np.sqrt((coincident_frame['a']*coincident_frame['t_2'])**2+\n",
" (coincident_frame['b']*coincident_frame['t_2'])**2+\n",
" (coincident_frame['c']*coincident_frame['t_2'])**2))\n",
"\n",
"## Check if it started inside and set the distance based on if it needs to travel to reach icecube or not\n",
"coincident_frame['Inside'] = (np.where((coincident_frame['t_1']/coincident_frame['t_2'] < 0) & (coincident_frame['z-position (m)'].between(-2450, -1450)), 1, 0))\n",
"coincident_frame['preliminary d'] = (np.where(coincident_frame['d_1'] <= coincident_frame['d_2'], coincident_frame['d_1'], coincident_frame['d_2']))\n",
"coincident_frame['d'] = (np.where(coincident_frame['Inside'] == 1, 0, coincident_frame['preliminary d']))\n",
"\n",
"## Check if the event lies in the cylinder\n",
"coincident_frame['In IC'] = (np.where((np.sqrt((coincident_frame['x-position (m)']-IceCube[station-1][0])**2 + (coincident_frame['y-position (m)']-IceCube[station-1][1])**2) < IceCube_radius) &\n",
" ((coincident_frame['z-position (m)']).between(-2450, -1450)) , 1, 0))\n",
"\n",
"#Correct coincident_frame to only have electron neutrinos inside IC\n",
"coincident_frame = coincident_frame[(((coincident_frame['In IC'] == 1) & (coincident_frame['flavor'] == 1)) | (coincident_frame['flavor'] == 2) | (coincident_frame['flavor'] == 3)) ]\n",
"\n",
"#Now calculate the lepton energies when they reach IC\n",
"coincident_frame['IC Mu Energies (eV)'] = (coincident_frame['Mu Energies (eV)'] * np.exp(-10**-5 * coincident_frame['d']*100)) # convert d from meters to cm\n",
"coincident_frame['weighted energies'] = (coincident_frame['weight'] * coincident_frame['Nu Energies (eV)'])"
]
},
{
"cell_type": "code",
"execution_count": 7,
"id": "f1757103",
"metadata": {
"scrolled": false
},
"outputs": [],
"source": [
"## Add possible Tau decay to the frame\n",
"coincident_frame['Tau decay'] = ''\n",
"# Again, the label 'Tau Decay' may be misleading because not all leptons may be taus\n",
"\n",
"## Calculate distance from the interaction point to its walls and keep the shortest (the first interaction with the volume)\n",
"\n",
"coincident_frame['distance-to-IC_1'] = np.sqrt((coincident_frame['x-positions'] - coincident_frame['x-intersect_1'])**2 + \n",
" (coincident_frame['y-positions'] - coincident_frame['y-intersect_1'])**2)\n",
"coincident_frame['distance-to-IC_2'] = np.sqrt((coincident_frame['x-positions'] - coincident_frame['x-intersect_2'])**2 + \n",
... 19001 more lines ...
|
| Attachment 4: IceCube_Relative_to_ARA_Stations.ipynb
|
{
"cells": [
{
"cell_type": "markdown",
"id": "a950b9af",
"metadata": {},
"source": [
"**This script is simply for me to calculate the location of IceCube relative to the origin of any ARA station**\n",
"\n",
"The relevant documentation to understand the definitions after the imports can be found in https://elog.phys.hawaii.edu/elog/ARA/130712_170712/doc.pdf"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "b926e2e3",
"metadata": {},
"outputs": [],
"source": [
"import numpy as np"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "901b442b",
"metadata": {},
"outputs": [],
"source": [
"#Definitions of translations in surveyor's coordinates:\n",
"\n",
"t_IC_to_ARAg = np.array([-24100, 1700, 6400])\n",
"t_ARAg_to_A1 = np.array([16401.71, -2835.37, -25.67])\n",
"t_ARAg_to_A2 = np.array([13126.7, -8519.62, -18.72])\n",
"t_ARAg_to_A3 = np.array([9848.35, -2835.19, -12.7])\n",
"\n",
"#Definitions of rotations from surveyor's axes to the ARA Station's coordinate systems\n",
"\n",
"R1 = np.array([[-0.598647, 0.801013, -0.000332979], [-0.801013, -0.598647, -0.000401329], \\\n",
" [-0.000520806, 0.0000264661, 1]])\n",
"R2 = np.array([[-0.598647, 0.801013, -0.000970507], [-0.801007, -0.598646,-0.00316072 ], \\\n",
" [-0.00311277, -0.00111477, 0.999995]])\n",
"R3 = np.array([[-0.598646, 0.801011, -0.00198193],[-0.801008, -0.598649,-0.00247504], \\\n",
" [-0.00316902, 0.000105871, 0.999995]])"
]
},
{
"cell_type": "markdown",
"id": "ab2d3206",
"metadata": {},
"source": [
"**Using these definitions, I should be able to calculate the location of IceCube relative to each ARA station by:**\n",
"\n",
"$$\n",
"\\vec{r}_{A 1}^{I C}=-R_1\\left(\\vec{t}_{I C}^{A R A}+\\vec{t}_{A R A}^{A 1}\\right)\n",
"$$\n",
"\n",
"We have a write-up of how to get this. Contact salcedogomez.1@osu.edu if you need that.\n",
"\n",
"Alex had done this already, he got that \n",
"\n",
"$$\n",
"\\vec{r}_{A 1}^{I C}=-3696.99^{\\prime} \\hat{x}-6843.56^{\\prime} \\hat{y}-6378.31^{\\prime} \\hat{z}\n",
"$$\n",
"\n",
"Let me verify that I get the same"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "912163d2",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A1 (in): [-3696.98956579 -6843.55800868 -6378.30926681]\n",
"IC coordinates relative to A1 (m): [-1127.13096518 -2086.4506124 -1944.60648378]\n",
"Distance of IC from A1 (m): 3066.788996234438\n"
]
}
],
"source": [
"IC_A1 = -R1 @ np.add(t_ARAg_to_A1, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A1 (in): \", IC_A1)\n",
"print(\"IC coordinates relative to A1 (m): \", IC_A1/3.28)\n",
"print(\"Distance of IC from A1 (m): \", np.sqrt((IC_A1[0]/3.28)**2 + (IC_A1[1]/3.28)**2 + (IC_A1[2]/3.28)**2))"
]
},
{
"cell_type": "markdown",
"id": "f9c9f252",
"metadata": {},
"source": [
"Looks good!\n",
"\n",
"Now, I just get the other ones:"
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "8afa27c6",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A2 (in): [ -1100.33577313 -12852.0589083 -6423.00776043]\n",
"IC coordinates relative to A2 (m): [ -335.46822352 -3918.31064277 -1958.2340733 ]\n",
"Distance of IC from A2 (m): 4393.219537890439\n"
]
}
],
"source": [
"IC_A2 = -R2 @ np.add(t_ARAg_to_A2, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A2 (in): \", IC_A2)\n",
"print(\"IC coordinates relative to A2 (m): \", IC_A2/3.28)\n",
"print(\"Distance of IC from A2 (m): \", np.sqrt((IC_A2[0]/3.28)**2 + (IC_A2[1]/3.28)**2 + (IC_A2[2]/3.28)**2))"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "9959d0a4",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A3 (in): [ -7609.73440732 -12079.45719852 -6432.31164368]\n",
"IC coordinates relative to A3 (m): [-2320.04097784 -3682.76134101 -1961.07062307]\n",
"Distance of IC from A3 (m): 4774.00452685144\n"
]
}
],
"source": [
"IC_A3 = -R3 @ np.add(t_ARAg_to_A3, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A3 (in): \", IC_A3)\n",
"print(\"IC coordinates relative to A3 (m): \", IC_A3/3.28)\n",
"print(\"Distance of IC from A3 (m): \", np.sqrt((IC_A3[0]/3.28)**2 + (IC_A3[1]/3.28)**2 + (IC_A3[2]/3.28)**2))"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "093dff67",
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.12"
}
},
"nbformat": 4,
"nbformat_minor": 5
}
|
|