| |
ID |
Date |
Author |
Subject |
Project |
|
|
31
|
Thu Dec 13 17:33:54 2018 |
s prohira | parallel jobs on ruby | Software | On ruby, users get charged for the full node, even if you aren't using all 20 cores, so it's a pain if you want to run a bunch of serial jobs. There is, however, a thing called the 'parallel command processor' (pcp) which is provided on ruby, (https://www.osc.edu/resources/available_software/software_list/parallel_command_processor) that makes it very simple.
essentially, you make a text file filled with commands, one command per line, and then you give it to the parallel command processor and it submits each line of your text file as an individual job. the nice thing about this is that you don't have to think about it. you just give it the file and go, and it will use all cores on the full node in the most efficient way possible.
below i provide 2 examples, a very simple one to show you how it works, and a more complicated one. in both files, i make the command file inside of a loop. you don't need to do this-you can make the file in some other way if you choose to. note that you can also do this from within an interactive job. more instructions at the above link.
test.pbs is just a minimal thing, where you need to submit the same command but with some value that needs to be incremented 1000 times (e.g. 1000 different jobs).
effvol.pbs is more involved, and shows some important steps if your job produces a lot of output, where you use the $TMPDIR or the pbs workdir. (if you don't know what that is, you probably don't need to use it). each command in this file stores an output file to the $TMPDIR directory. this directory is accessed faster than the directories where you store your files, and so your jobs run faster. at the end of the script, all of the output files from all of the run jobs, are copied to my home directory, because $TMPDIR is deleted after each job. also this file shows the sourcing of a particular bash profile for submitted jobs (if you need this. some programs work differently when submitted than jobs run on the login nodes on ruby).
i recommend reading the above link for more information. the pcp is very useful on ruby! |
| Attachment 1: test.pbs
|
#!/bin/bash
#PBS -A PCON0003
#PBS -l walltime=01:00:00
#PBS -l nodes=1:ppn=20
touch commandfile
for value in {1..1000}
do
line="/path/to/your_command_to_run $value (arg1) (arg2)..(argn)"
echo ${line}>> commandfile
done
module load pcp
mpiexec parallel-command-processor commandfile
|
| Attachment 2: effvol.pbs
|
#!/bin/bash
#PBS -A PCON0003
#PBS -N effvol
#PBS -l walltime=01:00:00
#PBS -l nodes=1:ppn=20
#PBS -o ./log/out
#PBS -e ./log/err
source /users/PCON0003/osu10643/.bash_batch
cd $TMPDIR
touch effvolconf
for value in {1..1000}
do
line="/users/PCON0003/osu10643/app/geant/app/nrt -m /users/PCON0003/osu10643/app/geant/app/nrt/effvol.mac -f \"$TMPDIR/effvol$value.root\""
echo ${line}>> effvolconf
done
module load pcp
mpiexec parallel-command-processor effvolconf
cp $TMPDIR/*summary.root /users/PCON0003/osu10643/doc/root/summary/
|
|
|
45
|
Fri Feb 4 13:06:25 2022 |
William Luszczak | "Help! AnitaBuildTools/PueoBuilder can't seem to find FFTW!" | Software | Disclaimer: This might not be the best solution to this problem. I arrived here after a lot of googling and stumbling across this thread with a similar problem for an unrelated project: https://github.com/xtensor-stack/xtensor-fftw/issues/52. If you're someone who actually knows cmake, maybe you have a better solution.
When compiling both pueoBuilder and anitaBuildTools, I have run into a cmake error that looks like:
CMake Error at /apps/cmake/3.17.2/share/cmake-3.17/Modules/FindPackageHandleStandardArgs.cmake:164 (message):
Could NOT find FFTW (missing: FFTW_LIBRARIES)
(potentially also missing FFTW_INCLUDES). Directing CMake to the pre-existing FFTW installations on OSC does not seem to do anything to resolve this error. From what I can tell, this might be related to how FFTW is built, so to get around this we need to build our own installation of FFTW using cmake instead of the recommended build process. To do this, grab the whatever version of FFTW you need from here: http://www.fftw.org/download.html (for example, I needed 3.3.9). Untar the source file into whatever directory you're working in:
tar -xzvf fftw-3.3.9.tar.gz
Then make a build directory and cd into it:
mkdir install
cd install
Now build using cmake, using the flags shown below.
cmake -DCMAKE_INSTALL_PREFIX=$(path_to_install_loc) -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON ../fftw-3.3.9
For example, I downloaded and untarred the source file in `/scratch/wluszczak/fftw/`, and my install prefix was `/scratch/wluszczak/fftw/install/`. In principle this installation prefix can be anywhere you have write access, but for the sake of organization I usually try to keep everything in one place.
Once you have configured cmake, go ahead and install:
make install -j $(nproc)
Where $(nproc) is the number of threads you want to use. On OSC I used $(nproc)=4 for compiling the ANITA tools and it finished in a reasonable amount of time.
Once this has finished, cd to your install directory and remove everything except the `include` and `lib64` folders:
cd $(path_to_install_dir) #You might already be here if you never left
rm *
rm -r CMakeFiles
Now we need to rebuild with slightly different flags:
cmake -DCMAKE_INSTALL_PREFIX=$(path_to_install_loc) -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON -DENABLE_FLOAT=ON ../fftw-3.3.9
make install -j $(nproc)
At the end of the day, your fftw install directory should have the following files:
include/fftw3.f
include/fftw3.f03
include/fftw3.h
include/fftw3l.f03
include/fftw3q.f03
lib64/libfftw3f.so
lib64/libfftw3f_threads.so.3
lib64/libfftw3_omp.so.3.6.9
lib64/libfftw3_threads.so
lib64/libfftw3f_omp.so
lib64/libfftw3f.so.3
lib64/libfftw3f_threads.so.3.6.9
lib64/libfftw3.so
lib64/libfftw3_threads.so.3
lib64/libfftw3f_omp.so.3
lib64/libfftw3f.so.3.6.9
lib64/libfftw3_omp.so
lib64/libfftw3.so.3
lib64/libfftw3_threads.so.3.6.9
lib64/libfftw3f_omp.so.3.6.9
lib64/libfftw3f_threads.so
lib64/libfftw3_omp.so.3
lib64/libfftw3.so.3.6.9
Once fftw has been installed, export your install directory (the one with the include and lib64 folders) to the following environment variable:
export FFTWDIR=$(path_to_install_loc)
Now you should be able to cd to your anitaBuildTools directory (or pueoBuilder directory) and run their associated build scripts:
./buildAnita.sh
or:
./pueoBuilder.sh
And hopefully your tools will magically compile (or at least, you'll get a new set of errors that are no longer related to this problem).
If you're running into an error that looks like:
CMake Error: The following variables are used in this project, but they are set to NOTFOUND.
Please set them or make sure they are set and tested correctly in the CMake files:
FFTWF_LIB (ADVANCED)
then pueoBuilder/anitaBuildTools can't seem to find your fftw installation (or files that are supposed to be included in that installation), try rebuilding FFTW with different flags according to which files it seems to think are missing.
If it seems like pueoBuilder can't seem to find your FFTW installation at all (i.e. you're getting some error that looks like missing: FFTW_LIBRARIES or missing: FFTW_INCLUDES, check the environment variables that are supposed to point to your local FFTW installation (`$FFTWDIR`) and make sure there are the correct files in the `lib` and `include` subdirectories. |
|
|
47
|
Mon Apr 24 11:51:42 2023 |
William Luszczak | PUEO simulation stack installation instructions | Software | These are instructions I put together as I was first figuring out how to compile PueoSim/NiceMC. This was originally done on machines running CentOS 7, however has since been replicated on the OSC machines (running RedHat 7.9 I think?). I generally try to avoid any `module load` type prerequisites, instead opting to compile any dependencies from source. You _might_ be able to get this to work by `module load`ing e.g. fftw, but try this at your own peril.
#pueoBuilder Installation Tutorial
This tutorial will guide you through the process of building the tools included in pueoBuilder from scratch, including the prerequisites and any environment variables that you will need to set. This sort of thing is always a bit of a nightmare process for me, so hopefully this guide can help you skip some of the frustration that I ran into. I did not have root acces on the system I was building on, so the instructions below are what I had to do to get things working with local installations. If you have root access, then things might be a bit easier. For reference I'm working on CentOS 7, other operating systems might have different problems that arise.
##Prerequisites
As far as I can tell, the prerequisites that need to be built first are:
-Python 3.9.18 (Apr. 6 2024 edit by Jason Yao, needed for ROOT 6.26-14)
-cmake 3.21.2 (I had problems with 3.11.4)
-gcc 11.1.0 (9.X will not work) (update 4/23/24: If you are trying to compile ROOT 6.30, you might need to downgrade to gcc 10.X, see note about TBB in "Issues I ran into" at the end)
-fftw 3.3.9
-gsl 2.7.1 (for ROOT)
-ROOT 6.24.00
-OneTBB 2021.12.0 (if trying to compile ROOT 6.30)
###CMake
You can download the source files for CMake here: https://cmake.org/download/. Untar the source files with:
tar -xzvf cmake-3.22.1.tar.gz
Compiling CMake is as easy as following the directions on the website: https://cmake.org/install/, but since we're doing a local build, we'll use the `configure` script instead of the listed `bootstrap` script. As an example, suppose that I downloaded the above tar file to `/scratch/wluszczak/cmake`:
mkdir install
cd cmake-3.22.1
./configure --prefix=/scratch/wluszczak/cmake/install
make
make install
You should additionally add this directory to your `$PATH` variable:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
To check to make sure that you are using the correct version of CMake, run:
cmake --version
and you should get:
cmake version 3.22.1
CMake suite maintained and supported by Kitware (kitware.com/cmake).
### gcc 11.1.0
Download the gcc source from github here: https://github.com/gcc-mirror/gcc/tags. I used the 11.1.0 release, though there is a more recent 11.2.0 release that I have not tried. Once you have downloaded the source files, untar the directory:
tar -xzvf gcc-releases-gcc-11.1.0.tar.gz
Then install the prerequisites for gcc:
cd gcc-releases-gcc-11.1.0
contrib/download_prerequisites
One of the guides I looked at also recommended installing flex separately, but I didn't seem to need to do this, and I'm not sure how you would go about it without root priviledges, though I imagine it's similar to the process for all the other packages here (download the source and then build by providing an installation prefix somewhere)
After you have installed the prerequisites, create a build directory:
cd ../
mkdir build
cd build
Then configure GCC for compilation like so:
../gcc-releases-gcc-11.1.0/configure -v --prefix=/home/wluszczak/gcc-11.1.0 --enable-checking=release --enable-languages=c,c++,fortran --disable-multilib --program-suffix=-11.1
I don't remember why I installed to my home directory instead of the /scratch/ directories used above. In principle the installation prefix can go wherever you have write access. Once things have configured, compile gcc with:
make -j $(nproc)
make install
Where `$(nproc)` is the number of processing threads you want to devote to compilation. More threads will run faster, but be more taxing on your computer. For reference, I used 8 threads and it took ~15 min to finish.
Once gcc is built, we need to set a few environment variables:
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
We also need to make sure cmake uses this compiler:
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/bin/gfortran-11.1
If your installation prefix in the configure command above was different, substitute that directory in place of `/home/wluszczak/gcc-11.1.0` for all the above export commands. To easily set these variables whenever you want to use gcc-11.1.0, you can stick these commands into a single shell script:
#load_gcc11.1.sh
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
(again substituting your installation prefix in place of mine). You can then set all these environment variables by simply running:
source load_gcc11.1.sh
Once this is done, you can check that gcc-11.1.0 is properly installed by running:
gcc-11.1 --version
Note that plain old
gcc --version
might still point to an older version of gcc. This is fine though.
###FFTW 3.3.9
Grab the source code for the appropriate versino of FFTW from here: http://www.fftw.org/download.html
However, do NOT follow the installation instructions on the webpage. Those instructions might work if you have root privileges, but I personally couldn't seem to to get things to work that way. Instead, we're going to build fftw with cmake. Untar the fftw source files:
tar -xzvf fftw-3.3.9.tar.gz
Make a build directory and cd into it:
mkdir build
cd build
Now build using cmake, using the flags shown below. For reference, I downloaded and untarred the source file in `/scratch/wluszczak/fftw/build`, so adjust your install prefix accordingly to point to your own build directory that you created in the previous step.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON ../fftw-3.3.9
make install -j $(nproc)
Now comes the weird part. Remove everything except the `include` and `lib64` directories in your build directory (if you installed to a different `CMAKE_INSTALL_PREFIX`, the include and lib64 directories might be located there instead. The important thing is that you want to remove everything, but leave the `include` and `lib64` directories untouched):
rm *
rm -r CMakeFiles
Now rebuild fftw, but with an additional flag:
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON -DENABLE_FLOAT=ON ../fftw-3.3.9
make install -j $(nproc)
At the end of the day, your fftw install directory should have the following files:
include/fftw3.f
include/fftw3.f03
include/fftw3.h
include/fftw3l.f03
include/fftw3q.f03
lib64/libfftw3f.so
lib64/libfftw3f_threads.so.3
lib64/libfftw3_omp.so.3.6.9
lib64/libfftw3_threads.so
lib64/libfftw3f_omp.so
lib64/libfftw3f.so.3
lib64/libfftw3f_threads.so.3.6.9
lib64/libfftw3.so
lib64/libfftw3_threads.so.3
lib64/libfftw3f_omp.so.3
lib64/libfftw3f.so.3.6.9
lib64/libfftw3_omp.so
lib64/libfftw3.so.3
lib64/libfftw3_threads.so.3.6.9
lib64/libfftw3f_omp.so.3.6.9
lib64/libfftw3f_threads.so
lib64/libfftw3_omp.so.3
lib64/libfftw3.so.3.6.9
Why do we have to do things this way? I don't know, I'm bad at computers. Maybe someone more knowledgeable knows. I found that when I didn't do this step, I'd run into errors that pueoBuilder could not find some subset of the required files (either the ones added by building with `-DENABLE_FLOAT`, or the ones added by building without `-DENABLE_FLOAT`).
Once fftw has been installed, export your install directory (the one with the include and lib64 folders) to the following environment variable:
export FFTWDIR=/scratch/wluszczak/fftw/build
Again, substituting your own fftw install prefix that you used above in place of `/scratch/wluszczak/fftw/build`
###gsl 2.7.1
gsl 2.7.1 is needed for the `mathmore` option in ROOT. If you have an outdated version of gsl, ROOT will still compile, but it will skip installing `mathmore` and `root-config --has-mathmore` will return `no`. To fix this, grab the latest source code for gsl from here: https://www.gnu.org/software/gsl/. Untar the files to a directory of your choosing:
tar -xzvf gsl-latest.tar.gz
For some reason I also installed gsl to my home directory, but in principle you can put it wherever you want.
mkdir /home/wluszczak/gsl
./configure --prefix=/home/wluszczak/gsl
make
make check
make install
To make sure ROOT can find this installation of gsl, you'll again need to set an environment variable prior to building ROOT:
export GSL_ROOT_DIR=/home/wluszczak/gsl/
I also added this to my $PATH variable, though I don't remember if that was required to get things working or not:
export PATH=/home/wluszczak/gsl/bin/:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gsl/lib:$LD_LIBRARY_PATH
###Python 3.9.18
Apr. 6, 2024 edit by Jason Yao:
I was able to follow this entire ELOG to install root 6.24.00, but I also was getting warnings/errors that seem to be related to Python,
so I went ahead and installed Python 3.9.18 (and then a newer version of ROOT just for fun).
Note that even though we can `module load python/3.9-2022.05` on OSC, I am 90% sure that this provided Python instance is no good as far as ROOT is concerned.
Head over to https://www.python.org/downloads/release/python-3918/ to check out the source code.
You can run
wget https://www.python.org/ftp/python/3.9.18/Python-3.9.18.tgz
to download it on OSC; then, run
tar -xzvf Python-3.9.18.tgz
cd Python-3.9.18
Next we will compile Python from source. I used this website for this step.
I wanted to install to `${HOME}/usr/Python-3.9.18/install`, so
./configure --prefix=${HOME}/usr/Python-3.9.18/install --enable-shared
Note that we must have the flag `--enable-shared` "to ensure that shared libraries are built for Python. By not doing this you are preventing any application which wants to use Python as an embedded environment from working" according to this guy.
(The corresponding error when you try to compile ROOT later on would look like "...can not be used when making a shared object; recompile with -fPIC...")
After configuration,
make -j8 && make install
(using 8 threads)
After installation, head over to the install directory, and then
cd bin
You should see `pip3` and `python3.9`. If you run
./python3
you should see the Python interactive terminal
Python 3.9.18 (main, Apr 5 2024, 22:49:51)
[GCC 11.1.0] on linux
Type "help", "copyright", "credits" or "license" for more information.
>>>
Add the python3 in this folder to your PATH variable. For example,
export PATH=${HOME}/usr/Python-3.9.18/install/bin:${PATH}
While we are in the python install directory, we might as well also use the `pip3` there to install numpy:
./pip3 install numpy
(I am not sure if this is absolutely needed by ROOT, but probably)
Next comes the important bit. You need to add the `lib/` directory inside you python installation to the environment variable $LD_LIBRARY_PATH
export LD_LIBRARY_PATH=${HOME}/usr/Python-3.9.18/install/lib:$LD_LIBRARY_PATH
according to stackoverflow. Without this step I ran into errors when compiling ROOT.
###ROOT 6.24.00
Download the specific version of ROOT that you need from here: https://root.cern/install/all_releases/
You might need to additionally install some of the dependencies (https://root.cern/install/dependencies/), but it seems like everything I needed was already installed on my system.
Untar the source you downloaded:
tar -xzvf root_v6.24.00.source.tar.gz
Make some build and install directories:
mkdir build install
cd build
Run CMake, but be sure to enable the fortan, mathmore and minuit2 options. For reference, I had downloaded and untarred the source files to `/scratch/wluszczak/root`. Your installation and source paths will be different.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/root/install/ /scratch/wluszczak/root/root-6.24.00/ -Dfortran=ON -Dminuit2=ON -Dmathmore=ON
Note: if you end up with an error related to compiling XROOTD, then add -Dxrootd=OFF to the original cmake command above.
Then proceed to start the build:
cmake --build . --target install -j $(nproc)
If everything has worked then after the above command finishes running, you should be able to run the following file to finish setting up ROOT:
source ../install/bin/thisroot.sh
##pueoBuilder
By this point, you should have working installations of CMake 3.21.2, gcc-11.1.0, fftw 3.3.9, and ROOT 6.24.00. Additionally, the following environment variables should have been set:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
export FFTWDIR=/scratch/wluszczak/fftw/build
At this point, the hard work is mostly done. Check out pueoBuilder with:
git clone git@github.com:PUEOCollaboration/pueoBuilder
set the following environment variables:
export PUEO_BUILD_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export PUEO_UTIL_INSTALL_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export NICEMC_SRC=${PUEO_BUILD_DIR}/components/nicemc
export NICEMC_BUILD=${PUEO_BUILD_DIR}/build/components/nicemc
export PUEOSIM_SRC=${PUEO_BUILD_DIR}/components/pueoSim
export LD_LIBRARY_PATH=${PUEO_UTIL_INSTALL_DIR}/lib:$LD_LIBRARY_PATH
Where $PUEO_BUILD_DIR and $PUEO_UTIL_INSTALL_DIR point to where you cloned pueoBuilder to (in my case, `/scratch/wluszczak/PUEO/pueoBuilder`. Now you should be able to just run:
./pueoBuilder.sh
Perform a prayer to the C++ gods while you're waiting for it to compile, and hopefully at the end of the day you'll have a working set of PUEO software.
##Issues I Ran Into
If you already have an existing installation of ROOT, you may still need to recompile to make sure you're using the same c++ standard that the PUEO software is using. I believe the pre-compiled ROOT binaries available through their website are insufficient, though maybe someone else has been able to get those working.
If you're running into errors about c++ standard or compiler version even after you have installed gcc-11.1.0, then for some reason your system isn't recognizing your local installation of gcc-11.1.0. Check the path variables ($PATH and $LD_LIBRARY_PATH) to make sure the gcc-11.1.0 `bin` directory is being searched.
If you're running into an error that looks like:
CMake Error: The following variables are used in this project, but they are set to NOTFOUND.
Please set them or make sure they are set and tested correctly in the CMake files:
FFTWF_LIB (ADVANCED)
then pueoBuilder can't seem to find your fftw installation (or files that are supposed to be included in that installation), try rebuilding with different flags according to which files it seems to think are missing.
If it seems like pueoBuilder can't seem to find your fftw installation at all (i.e. you're getting some error that looks like `missing: FFTW_LIBRARIES` or `missing: FFTW_INCLUDES`), check the environment variables that are supposed to point to your fftw installation (`$FFTWDIR`) and make sure there are the correct files in the `lib` and `include` subdirectories.
Update: 6/23/24: The latest version of ROOT (6.30) will fail to compile on OSC unless you manually compile TBB as well. An easy workaround is to simply downgrade to ROOT 6.24, however if you really need ROOT 6.30 you can follow the instructions below to install TBB and compile ROOT:
You will first need to downgrade to GCC 10.X. TBB will not compile with GCC 11. This can be done by following the GCC installation isntructions above, except starting with GCC 10 source code instead of GCC 11.
To install TBB yourself, download the source code (preferably the .tar.gz file) from here: https://github.com/oneapi-src/oneTBB/releases/tag/v2021.12.0. Move the file to the directory where you want to install TBB and untar it with:
tar -xzvf oneTBB-2021.12.0.tar.gz
Make some build and install directories:
mkdir build install
cd build
Then configure cmake:
cmake -DCMAKE_INSTALL_PREFIX=/path/to/tbb/install
Then compile with:
cmake --build .
Once this has finished running, you can add the installation to you $PATH and $LD_LIBRARY_PATH variables:
export PATH=/path/to/tbb/install/bin:$PATH
export LD_LIBRARY_PATH=/path/to/tbb/install/lib64:$LD_LIBRARY_PATH
You can then proceed as normal, except when compiling root you will need one additional cmake flag (Dbuiltin_tbb=ON):
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/root/install/ /scratch/wluszczak/root/root-6.24.00/ -Dfortran=ON -Dminuit2=ON -Dmathmore=ON -Dbuiltin_tbb=ON
And hopefully this should work. This process is a little bit more involved than just downgrading ROOT, so try to avoid going down this route unless absolutely necessary.
|
|
|
Draft
|
Sun Sep 17 20:05:29 2017 |
Spoorthi Nagasamudram | Some basic | | |
|
|
Draft
|
Thu Apr 27 18:28:22 2017 |
Sam Stafford (Also Slightly Jacob) | Installing AnitaTools on OSC | Software | Jacob Here, Just want to add how I got AnitaTools to see FFTW:
1) echo $FFTW3_HOME to find where the lib and include dir is.
2) Next add the following line to the start of cmake/modules/FindFFTW.cmake
'set ( FFTW_ROOT full/path/you/got/from/step/1)'
Brief, experience-based instructions on installing the AnitaTools package on the Oakley OSC cluster. |
| Attachment 1: OSC_build.txt
|
Installing AnitaTools on OSC
Sam Stafford
04/27/2017
This document summarizes the issues I encountered installing AnitaTools on the OSC Oakley cluster.
I have indicated work-arounds I made for unexpected issues
I do not know that this is the only valid process
This process was developed by trial-and-error (mostly error) and may contain superfluous steps
A person familiar with AnitaTools and cmake may be able to streamline it
Check out OSC's web site, particularly to find out about MODULES, which facilitate access to pre-installed software
export the following environment variables in your .bash_profile (not .bashrc):
ROOTSYS where you want ROOT to live
install it in somewhere your user directory; at this time, ROOT is not pre-installed on Oakley as far as I can tell
ANITA_UTIL_INSTALL_DIR where you want anitaTools to live
FFTWDIR where fftw is
look on OSC's website to find out where it is; you shouldn't have to install it locally
PATH should contain $FFTWDIR/bin and $ROOTSYS/bin
LD_LIBRARY_PATH should contain $FFTWDIR/lib $ROOTSYS/lib $ANITA_UTIL_INSTALL_DIR/lib
LD_INCLUDE_PATH should contain $FFTWDIR/include $ROOTSYS/include $ANITA_UTIL_INSTALL_DIR/include
also put in your .bash_profile: (I put these after the exports)
module load gnu/4.8.5 // loads g++ compiler
(this should automatically load module fftw/3.3.4 also)
install ROOT - follow ROOT's instructions to build from source. It's a typical (configure / make / make install) sequence
you probably need ./configure --enable-Minuit2
get AnitaTools from github/anitaNeutrino "anitaBuildTool" (see Anita ELOG 672, by Cosmin Deaconu)
Change entry in which_event_reader to ANITA3, if you want to analyze ANITA-3 data
(at least for now; I think they are developing smarts to make the SW adapt automatically to the anita data "version")
Do ./buildAnita.sh //downloads the software and attempts a full build/install
it may file on can't-find-fftw during configure:
system fails to populate environment variable FFTW_ROOT, not sure why
add the following line at beginning of anitaBuildTool/cmake/modules/FindFFTW.cmake:
set( FFTW_ROOT /usr/local/fftw3/3.3.4-gnu)
(this apparently tricks cmake into finding fftw)
NOTE: ./buildAnita.sh always downloads the software from github. IT WILL WIPE OUT ANY CHANGES YOU MADE TO AnitaTools!
Do "make"
May fail with /usr/lib64/libstdc++.so.6: version `GLIBCXX_3.4.14' not found and/or a few other similar messages
or may say c11 is not supported
need to change compiler setting for cmake:
make the following change in anitaBuildTool/build/CMakeCache.txt (points cmake to the g++ compiler instead of default intel/c++)
#CMAKE_CXX_COMPILER:FILEPATH=/usr/bin/c++ (comment this out)
CMAKE_CXX_COMPILER:FILEPATH=/usr/local/gcc/4.8.5/bin/g++ (add this)
(you don't necessarfily have to use version, gcc/4.8.5, but it worked for me)
Then retry by doing "make"
Once make is completed, do "make install"
A couple of notes:
Once AnitaTools is built, if you change source, just do make, then make install (from anitaBuildTool) (don't ./buildAnita.sh; see above)
(actually make install will do the make step if source changes are detected)
To start AnitaTools over from cmake, delete the anitaBuildTool/build directory and run make (not cmake: make will drive cmake for you)
(don't do cmake directly unless you know what you're doing; it'll mess things up)
|
|
|
9
|
Thu May 11 13:43:46 2017 |
Sam Stafford | Notes on installing icemc on OSC | Software | |
| Attachment 1: icemc_setup_osc.txt
|
A few notes about installing icemc on OSC
Dependencies
ROOT - download from CERN and install according to instructions
FFTW - do "module load gnu/4.8.5" (or put it in your .bash_profile)
The environment variable FFTWDIR must contain the directory where FFTW resides
in my case this was /usr/local/fftw3/3.3.4-gnu
set this up in your .bash_profile (not .bashrc)
I copied my working instance of icemc from my laptop to a folder in my osc space
Copy the whole icemc directory (maybe its icemc/trunk, depending on how you installed), EXCEPT for the "output" subdir because it's big and unnecessary
in your icemc directory on OSC, do "mkdir output"
In icemc/Makefile
find a statement like this:
LIBS += -lMathMore $(FFTLIBS) -lAnitaEvent
and modify it to include the directory where the FFTW library is:
LIBS += -L$(FFTWDIR)/lib -lMathMore $(FFTLIBS) -lAnitaEvent
note: FFTLIBS contains the list of libraries (e.g., -lfftw3), NOT the library search paths
Compile by doing "make"
Remember you should set up a batch job on OSC using PBS.
|
|
|
10
|
Thu May 11 14:38:10 2017 |
Sam Stafford | Sample OSC batch job setup | Software | Batch jobs on OSC are initiated through the Portable Batch System (PBS). This is the recommended way to run stuff on OSC clusters.
Attached is a sample PBS script that copies files to temporary storage on the OSC cluster (also recommended) and runs an analysis program.
Info on batch processing is at https://www.osc.edu/supercomputing/batch-processing-at-osc.
This will tell you how to submit and manage batch jobs.
More resources are available at www.osc.edu.
PBS web site: /www.pbsworks.com
The PBS user manual is at www.pbsworks.com/documentation/support/PBSProUserGuide10.4.pdf. |
| Attachment 1: osc_batch_jobs.txt
|
## annotated sample PBS batch job specification for OSC
## Sam Stafford 05/11/2017
#PBS -N j_ai06_${RUN_NUMBER}
##PBS -m abe ## request an email on job completion
#PBS -l mem=16GB ## request 16GB memory
##PBS -l walltime=06:00:00 ## set this in qsub
#PBS -j oe ## merge stdout and stderr into a single output log file
#PBS -A PAS0174
echo "run number " $RUN_NUMBER
echo "cal pulser " $CAL_PULSER
echo "baseline file " $BASELINE_FILE
echo "temp dir is " $TMPDIR
echo "ANITA_DATA_REMOTE_DIR="$ANITA_DATA_REMOTE_DIR
set -x
## copy the files from kingbee to the temporary workspace
## (if you set up public key authentication between kingbee and OSC, you won't need a password; just google "public key authentication")
mkdir $TMPDIR/run${RUN_NUMBER} ## make a directory for this run number
scp stafford.16@kingbee.mps.ohio-state.edu:/data/anita/anita3/flightData/copiedBySam/run${RUN_NUMBER}/calEventFile${RUN_NUMBER}.root $TMPDIR/run${RUN_NUMBER}/calEventFile${RUN_NUMBER}.root
scp stafford.16@kingbee.mps.ohio-state.edu:/data/anita/anita3/flightData/copiedBySam/newerData/run${RUN_NUMBER}/gpsEvent${RUN_NUMBER}.root $TMPDIR/run${RUN_NUMBER}/gpsEvent${RUN_NUMBER}.root
scp stafford.16@kingbee.mps.ohio-state.edu:/data/anita/anita3/flightData/copiedBySam/newerData/run${RUN_NUMBER}/timedHeadFile${RUN_NUMBER}.root $TMPDIR/run${RUN_NUMBER}/timedHeadFile${RUN_NUMBER}.root
scp stafford.16@kingbee.mps.ohio-state.edu:/data/anita/anita3/flightData/copiedBySam/newerData/run${RUN_NUMBER}/decBlindHeadFileV1_${RUN_NUMBER}.root $TMPDIR/run${RUN_NUMBER}/decBlindHeadFileV1_${RUN_NUMBER}.root
## set up the environment variables to point to the temporary work space
export ANITA_DATA_REMOTE_DIR=$TMPDIR
export ANITA_DATA_LOCAL_DIR=$TMPDIR
echo "ANITA_DATA_REMOTE_DIR="$ANITA_DATA_REMOTE_DIR
## run the analysis program
cd analysisSoftware
./analyzerIterator06 ${CAL_PULSER} -S1 -Noverlap --FILTER_OPTION=4 ${BASELINE_FILE} ${RUN_NUMBER} -O
echo "batch job ending"
|
|
|
14
|
Mon Sep 18 12:06:01 2017 |
Oindree Banerjee | How to get anitaBuildTool and icemc set up and working | Software | First try reading and following the instructions here
https://u.osu.edu/icemc/new-members-readme/
Then e-mail me at oindreeb@gmail.com with your problems
|
|
|
35
|
Tue Feb 26 19:07:40 2019 |
Lauren Ennesser | Valgrind command to suppress ROOT warnings | | valgrind --suppressions=$ROOTSYS/etc/valgrind-root.supp ./myCode
If you use valgrind to identify potential memory leaks in your code, but use a lot of ROOT objects and functions, you'll notice that ROOTs TObjects trigger a lot of "potential memory leak" warnings. This option will suppress many of those. More info at https://root-forum.cern.ch/t/valgrind-and-root/2ss8506 |
|
|
42
|
Tue Nov 5 16:22:16 2019 |
Keith McBride | NASA Proposal fellowships | Other | Here is a useful link for astroparticle grad students related proposals from NASA:
https://nspires.nasaprs.com/external/solicitations/summary.do?solId=%7BE16CD59F-29DD-06C0-8971-CE1A9C252FD4%7D&path=&method=init
Full email I received regarding this information was:
_______________________________________________________________________________________________________________________________________________________________________________________________________________________________________________
ROSES-19 Amendment adds a new program element to ROSES-2019: Future Investigators in NASA Earth and Space Science and Technology (FINESST), the ROSES Graduate Student Research Program Element, E.6.
Through FINESST, the Science Mission Directorate (SMD) solicits proposals from accredited U.S. universities and other eligible organizations for graduate student-designed and performed research projects that contribute to SMD's science, technology and exploration goals.
A Notice of Intent is not requested for E.6 FINESST. Proposals to FINESST are due by February 4, 2020.
Potential proposers who wish to participate in the optional pre-proposal teleconference December 2, 2019 from 1:00-2:30 p.m. Eastern Time may (no earlier than 30 minutes prior to the start time) call 1-888-324-3185 (U.S.-only Toll Free) or 1-630-395-0272 (U.S. Toll) and use Participant Passcode: 8018549. Restrictions may prevent the use of a toll-free number from a mobile or free-phone or from telephones outside the U.S. For U.S. TTY-equipped callers or other types of relay service no earlier than 30 minutes before the start of the teleconference, call 711 and provide the same conference call number/passcode. Email HQ-FINESST@mail.nasa.gov any teleconference agenda suggestions and questions by November 25, 2019. Afterwards, questions and responses, with identifying information removed, will be posted on the NSPIRES page for FINESST under "other documents".
FINESST awards research grants with a research mentor as the principal investigator and the listed graduate student listed as the "student participant". Unlike the extinct NASA Earth and Space Science Fellowships (NESSF), the Future Investigators (FIs) are not trainees or fellows. Students with existing NESSF awards seeking a third year of funding, may not submit proposals to FINESST. Instead, they may propose to NESSF20R. Subject to a period of performance restriction, some former NESSFs may be eligible to submit proposals to FINESST.
On or about November 1, 2019, this Amendment to the NASA Research Announcement "Research Opportunities in Space and Earth Sciences (ROSES) 2019" (NNH19ZDA001N) will be posted on the NASA research opportunity homepage at http://solicitation.nasaprs.com/ROSES2019 and will appear on the RSS feed at: https://science.nasa.gov/researchers/sara/grant-solicitations/roses-2019/
Questions concerning this program element may be directed to HQ-FINESST@mail.nasa.gov. |
|
|
39
|
Thu Jul 11 10:05:37 2019 |
Justin Flaherty | Installing PyROOT for Python 3 on Owens | Software | In order to get PyROOT working for Python 3, you must build ROOT with a flag that specifies Python 3 in the installation. This method will create a folder titled root-6.16.00 in your current directory, so organize things how you see fit. Then the steps are relatively simple:
wget https://root.cern/download/root_v6.16.00.source.tar.gz
tar -zxf root_v6.16.00.source.tar.gz
cd root-6.16.00
mkdir obj
cd obj
cmake .. -Dminuit2=On -Dpython3=On
make -j8
If you wish to do a different version of ROOT, the steps should be the same:
wget https://root.cern/download/root_v<version>.source.tar.gz
tar -zxf root_v<version>.source.tar.gz
cd root-<version>
mkdir obj
cd obj
cmake .. -Dminuit2=On -Dpython3=On
make -j8 |
|
|
Draft
|
Fri Feb 28 13:09:53 2020 |
Justin Flaherty | Installing anitaBuildTools on OSC-Owens (Revised 2/28/2020) | | |
|
|
Draft
|
Thu Sep 21 14:30:18 2017 |
Julie Rolla | Using/Running XF | | Below I've attached a video with some information regarding running XF. Before you start, here's some important information you need to know.
In order to run XF:
XF can now only run on a windows OS. If you are on a Mac, you can dual boot with windows 10 that is not activated—this still works. The lack of activation gives you minimal contraints, and will not hinder your ability to run XF. Otherwise, you can use a queen bee machine. Note that there are two ways to run XF itself (once you are on a windows machine). You can run via OSC on Oakly--This has a floating license and you will not need to use the USB Key-- or you can use the USB key. Unfortunately, at the moment, we only have one USB key, and it may be best to get an account through OSC.
2 methods:
1.) If you are not using Oakly— you need the USB key to run
2.) Log in to Oakly — you do not need the USB key to run
------
for method 1:
XFdtd you can just click on the icon
Will pop up with “need to find a valid license”
click “ok” and insert the USB key and hit “retry"
For method 2:
Log in to Oakly — To log into Oakly follow the steps between **
** ssh in
put “module load XFdtd” in command line— this gets you access to the libraries
then put “XFdtd” to load it
————
Note: you must have drivers installed to use usb key. Once you plug it in the first time, it will tell you what to install.
Note: The genetic algorithm will change the geometry of the detector and XF will check the gain values with those given geometries.
After XF is loaded:
Components are listed on the left. Each are different for different simulations.
To put in geometry on antenna, click on “parts”.
click “create new”
choose a type of geometry.
Note that you can also go to “file” and import and you can import cad files.
“Create simulation”— save the project, and give it a name. Then, click “create simulation”. This stores all of the geometry and settings of the simulation. Now you could, if you wanted to, browse all of the different types of simulations.
How to actually run the simulations:
In this example, Carl setup a planar sensor with a point sensor inside the sphere, and two more sensors on each side of the sphere. Now, you load the simulation and hit the play button at the bottom. Note that this should take 20 or 30 minutes for it to actually simulate. When it is done, you can actually re-click the play button and it will show you the visual simulation. It will automatically write this out when running the simulations. You either now need to parse that, or be able to view the data in XF itself.
You can click “file” ad export data. Additionally, you can export the image. Note that this can give you a “smith chart”; this is that gain measurement you’re looking for. If you had a far field/zone sensor, then you could get a far field gain— which is this smith chart. To get the smith chart, you hover over the sensor, and right click. This should give you the option of a smith chart if you had the correct sensor. Note that all of this data and information is on the right hand side under “results” this will pull up all of the sensors, which you can right click to gather the actual individual data on.
Note: the far zone sensor puts sensors completely symmetrical around the object. Ie if we have a sphere, we will have a larger sphere outside of our conducting sphere/antenna. |
|
|
34
|
Tue Feb 26 16:19:20 2019 |
Julie Rolla | All of the group GitHub account links | Software | ANITA Binned Analysis: https://github.com/osu-particle-astrophysics/BinnedAnalysis
GENETIS Bicone: https://github.com/mclowdus/BiconeEvolution
GENETIS Dipole: https://github.com/hchasan/XF-Scripts
ANITA Build tool: https://github.com/anitaNeutrino/anitaBuildTool
ANITA Hackathon: https://github.com/anitaNeutrino/hackathon2017
ICEMC: https://github.com/anitaNeutrino/icemc
Brian's Github: https://github.com/clark2668?tab=repositories
Note that you *may* need permissions for some of these. Please email Lauren (ennesser.1@buckeyemail.osu.edu ), Julie (JulieRolla@gmail.com), AND Brian (clark.2668@buckeyemail.osu.edu ) if you have any issues with permssions. Please state which GitHub links you are looking to view. |
|
|
6
|
Tue Apr 25 10:22:50 2017 |
Jude Rajasekera | ShelfMC Cluster Runs | Software | Doing large runs of ShelfMC can be time intensive. However, if you have access to a computing cluster like Ruby or KingBee, where you are given a node with multiple processors, ShelfMC runs can be optimized by utilizing all available processors on a node. The multithread_shelfmc.sh script automates these runs for you. The script and instructions are attached below. |
| Attachment 1: multithread_shelfmc.sh
|
#!/bin/bash
#Jude Rajasekera 3/20/2017
shelfmcDir=/users/PCON0003/cond0091/ShelfMC #put your shelfmc directory address here
runName='TestRun' #name of run
NNU=500000 #total NNU per run
seed=42 #initial seed for every run,each processor will recieve a different seed (42,43,44,45...)
NNU="$(($NNU / 20))" #calculating NNU per processor, change 20 to however many processor your cluster has per node
ppn=20 #processors per node
########################### make changes for input.txt file here #####################################################
input1="#inputs for ARIANNA simulation, do not change order unless you change ReadInput()"
input2="$NNU #NNU, setting to 1 for unique neutrino"
input3="$seed #seed Seed for Rand3"
input4="18.0 #EXPONENT, !should be exclusive with SPECTRUM"
input5="1000 #ATGap, m, distance between stations"
input6="4 #ST_TYPE, !restrict to 4 now!"
input7="4 #N_Ant_perST, not to be confused with ST_TYPE above"
input8="2 #N_Ant_Trigger, this is the minimum number of AT to trigger"
input9="30 #Z for ST_TYPE=2"
input10="575 #ICETHICK, thickness of ice including firn, 575m at Moore's Bay"
input11="1 #FIRN, KD: ensure DEPTH_DEPENDENT is off if FIRN is 0"
input12="1.30 #NFIRN 1.30"
input13="$122 #FIRNDEPTH in meters"
input14="1 #NROWS 12 initially, set to 3 for HEXAGONAL"
input15="1 #NCOLS 12 initially, set to 5 for HEXAGONAL"
input16="0 #SCATTER"
input17="1 #SCATTER_WIDTH,how many times wider after scattering"
input18="0 #SPECTRUM, use spectrum, ! was 1 initially!"
input19="0 #DIPOLE, add a dipole to the station, useful for st_type=0 and 2"
input20="0 #CONST_ATTENLENGTH, use constant attenuation length if ==1"
input21="1000 #ATTEN_UP, this is the conjuction of the plot attenlength_up and attlength_down when setting REFLECT_RATE=0.5(3dB)"
input22="250 #ATTEN_DOWN, this is the average attenlength_down before Minna Bluff measurement(not used anymore except for CONST_ATTENLENGTH)"
input23="4 #NSIGMA, threshold of trigger"
input24="1 #ATTEN_FACTOR, change of the attenuation length"
input25="1 #REFLECT_RATE,power reflection rate at the ice bottom"
input26="0 #GZK, 1 means using GZK flux, 0 means E-2 flux"
input27="0 #FANFLUX, use fenfang's flux which only covers from 10^17 eV to 10^20 eV"
input28="0 #WIDESPECTRUM, use 10^16 eV to 10^21.5 eV as the energy spectrum, otherwise use 17-20"
input29="1 #SHADOWING"
input30="1 #DEPTH_DEPENDENT_N;0 means uniform firn, 1 means n_firn is a function of depth"
input31="0 #HEXAGONAL"
input32="1 #SIGNAL_FLUCT 1=add noise fluctuation to signal or 0=do not"
input33="4.0 #GAINV gain dependency"
input34="1 #TAUREGENERATION if 1=tau regeneration effect, if 0=original"
input35="3.0 #ST4_R radius in meters between center of station and antenna"
input36="350 #TNOISE noise temperature in Kelvin"
input37="80 #FREQ_LOW low frequency of LPDA Response MHz #was 100"
input38="1000 #FREQ_HIGH high frequency of LPDA Response MHz"
input39="/users/PCON0003/cond0091/ShelfMC/temp/LP_gain_manual.txt #GAINFILENAME"
###########################################################################################
cd $shelfmcDir #cd to dir containing shelfmc
mkdir $runName #make a folder for run
cd $runName #cd into run folder
initSeed=$seed
for (( i=1; i<=$ppn;i++)) #make 20 setup files for 20 processors
do
mkdir Setup$i #make setup folder i
cd Setup$i #go into setup folder i
seed="$(($initSeed+$i-1))" #calculate seed for this iteration
input3="$seed #seed Seed for Rand3" #save new input line
for j in {1..40} #print all input.txt lines
do
lineName=input$j
echo "${!lineName}" >> input.txt #print line to input.txt file
done
cd ..
done
pwd=`pwd`
#create job file
echo '#!/bin/bash' >> run_shelfmc_multithread.sh
echo '#PBS -l nodes=1:ppn='$ppn >> run_shelfmc_multithread.sh #change depending on processors per node
echo '#PBS -l walltime=00:05:00' >> run_shelfmc_multithread.sh #change walltime depending on run size, will be 20x shorter than single processor run time
echo '#PBS -N shelfmc_'$runName'_job' >> run_shelfmc_multithread.sh
echo '#PBS -j oe' >> run_shelfmc_multithread.sh
echo '#PBS -A PCON0003' >> run_shelfmc_multithread.sh #change to specify group
echo 'cd ' $shelfmcDir >> run_shelfmc_multithread.sh
echo 'runName='$runName >> run_shelfmc_multithread.sh
for (( k=1; k<=$ppn;k++))
do
echo './shelfmc_stripped.exe $runName/Setup'$k' _'$k'$runName &' >> run_shelfmc_multithread.sh #execute commands for 20 setup files
done
echo 'wait' >> run_shelfmc_multithread.sh #wait until all runs are finished
echo 'cd $runName' >> run_shelfmc_multithread.sh #go into run folder
echo 'for (( i=1; i<='$ppn';i++)) #20 iterations' >> run_shelfmc_multithread.sh
echo 'do' >> run_shelfmc_multithread.sh
echo ' cd Setup$i #cd into setup dir' >> run_shelfmc_multithread.sh
echo ' mv *.root ..' >> run_shelfmc_multithread.sh #move root files to runDir
echo ' cd ..' >> run_shelfmc_multithread.sh
echo 'done' >> run_shelfmc_multithread.sh
echo 'hadd Result_'$runName'.root *.root' >> run_shelfmc_multithread.sh #add all root files
echo 'rm *ShelfMCTrees*' >> run_shelfmc_multithread.sh #delete all partial root files
chmod u+x run_shelfmc_multithread.sh
echo "Run files created"
echo "cd into run folder and do $ qsub run_shelfmc_multithread.sh"
|
| Attachment 2: multithread_shelfmc_walkthrough.txt
|
This document will explain how to dowload, configure, and run multithread_shelfmc.sh in order to do large runs on computing clusters.
####DOWNLOAD####
1.Download multithread_shelfmc.sh
2.Move multithread_shelfmc.sh into ShelfMC directory
3.Do $chmod u+x multithread_shelfmc.sh
####CONFIGURE###
1.Open multithread_shelfmc.sh
2.On line 3, modify shelfmcDir to your ShelfMC dir
3.On line 6, add your run name
4.On line 7, add the total NNU
5.On line 8, add an intial seed
6.On line 10, specify number of processors per node for your cluster
7.On lines 12-49, edit the input.txt parameters
8.On line 50, add the location of your LP_gain_manual.txt
9.On line 80, specify a wall time for each run, remember this will be about 20x shorter than ShelfMC on a single processor
10.On line 83, Specify the group name for your cluster if needed
11.Save file
####RUN####
1.Do $./multithread_shelfmc.sh
2.There should now be a new directory in the ShelfMC dir with 20 setup files and a run_shelfmc_multithread.sh script
3.Do $qsub run_shelfmc_multithread.sh
###RESULT####
1.After the run has completed, there will be a result .root file in the run directory
|
|
|
7
|
Tue Apr 25 10:35:43 2017 |
Jude Rajasekera | ShelfMC Parameter Space Scan | Software | These scripts allow you to do thousands of ShelfMC runs while varying certain parameters of your choice. As is, the attenuation length, reflection, ice thickness, firn depth, station depth is varied over certain rages; in total, the whole Parameter Space Scan does 5250 runs on a cluster like Ruby or KingBee. The scripts and instructions are attached below. |
| Attachment 1: ParameterSpaceScan_instructions.txt
|
This document will explain how to dowload, configure, and run a parameter space search for ShelfMC on a computing cluster.
These scripts explore the ShelfMC parameter space by varying ATTEN_UP, REFLECT_RATE, ICETHICK, FIRNDEPTH, and STATION_DEPTH for certain rages.
The ranges and increments can be found in setup.sh.
In order to vary STATION_DEPTH, some changes were made to the ShelfMC code. Follow these steps to allow STATION_DEPTH to be an input parameter.
1.cd to ShelfMC directory
2.Do $sed -i -e 's/ATDepth/STATION_DEPTH/g' *.cc
3.Open declaration.hh. Replace line 87 "const double ATDepth = 0.;" with "double STATION_DEPTH;"
4.In functions.cc go to line 1829. This is the ReadInput() method. Add the lines below to the end of this method.
GetNextNumber(inputfile, number); // new line for station Depth
STATION_DEPTH = (double) atof(number.c_str()); //new line
5.Do $make clean all
#######Script Descriptions########
setup.sh -> This script sets up the necessary directories and setup files for all the runs
scheduler.sh -> This script submits and monitors all jobs.
#######DOWNLOAD########
1.Download setup.sh and scheduler.sh
2.Move both files into your ShelfMC directory
3.Do $chmod u+x setup.sh and $chmod u+x scheduler.sh
######CONFIGURE#######
1.Open setup.sh
2.On line 4, modify the job name
3.On line 6, modify group name
4.On line 10, specify your ShelfMC directory
5.On line 13, modify your run name
6.On line 14, specify the NNU per run
7.On line 15, specify the starting seed
8.On line 17, specify the number of processors per node on your cluster
9.On lines 19-56, edit the input.txt parameters that you want to keep constant for every run
10.On line 57, specify the location of the LP_gain_manual.txt
11.On line 126, change walltime depending on total NNU. Remember this wall time will be 20x shorter than a single processor run.
12.On line 127, change job prefix
13.On line 129, change the group name if needed
14.Save file
15.Open scheduler.sh
16.On line 4, specify your ShelfMC directory
17.On line 5, modify run name. Make sure it is the same runName as you have in setup.sh
18.On lines 35 and 39, replace cond0091 with your username for the cluster
19.On line 42, you can pick how many nodes you want to use at any given time. It is set to 6 intially.
20.Save file
#######RUN#######
1.Do $qsub setup.sh
2.Wait for setup.sh to finish. This script is creating the setup files for all runs. This may take about an hour.
3.When setup.sh is done, there should be a new directory in your home directory. Move this directory to your ShelfMC directory.
4.Do $screen to start a new screen that the scheduler can run on. This is incase you lose connection to the cluster mid run.
5.Do $./scheduler.sh to start script. This script automatically submits jobs and lets you see the status of the runs. This will run for several hours.
5.The scheduler makes a text file of all jobs called jobList.txt in the ShelfMC dir. Make sure to delete jobList.txt before starting a whole new run.
######RESULT#######
1.When Completed, there will be a great amount of data in the run files, about 460GB.
2.The run directory is organized in tree, results for particular runs can be found by cd'ing deeper into the tree.
3.In each run directory, there will be a resulting root file, all the setup files, and a log file for the run.
|
| Attachment 2: setup.sh
|
#!/bin/bash
#PBS -l walltime=04:00:00
#PBS -l nodes=1:ppn=1,mem=4000mb
#PBS -N jude_SetupJob
#PBS -j oe
#PBS -A PCON0003
#Jude Rajasekera 3/20/17
#directories
WorkDir=$TMPDIR
tmpShelfmc=$HOME/shelfmc/ShelfMC #set your ShelfMC directory here
#controlled variables for run
runName='ParamSpaceScanDir' #name of run
NNU=500000 #NNU per run
seed=42 #starting seed for every run, each processor will recieve a different seed (42,43,44,45...)
NNU="$(($NNU / 20))" #calculating processors per node, change 20 to however many processors your cluster has per node
ppn=5 #number of processors per node on cluster
########################### input.txt file ####################################################
input1="#inputs for ARIANNA simulation, do not change order unless you change ReadInput()"
input2="$NNU #NNU, setting to 1 for unique neutrino"
input3="$seed #seed Seed for Rand3"
input4="18.0 #EXPONENT, !should be exclusive with SPECTRUM"
input5="1000 #ATGap, m, distance between stations"
input6="4 #ST_TYPE, !restrict to 4 now!"
input7="4 #N_Ant_perST, not to be confused with ST_TYPE above"
input8="2 #N_Ant_Trigger, this is the minimum number of AT to trigger"
input9="30 #Z for ST_TYPE=2"
input10="$T #ICETHICK, thickness of ice including firn, 575m at Moore's Bay"
input11="1 #FIRN, KD: ensure DEPTH_DEPENDENT is off if FIRN is 0"
input12="1.30 #NFIRN 1.30"
input13="$FT #FIRNDEPTH in meters"
input14="1 #NROWS 12 initially, set to 3 for HEXAGONAL"
input15="1 #NCOLS 12 initially, set to 5 for HEXAGONAL"
input16="0 #SCATTER"
input17="1 #SCATTER_WIDTH,how many times wider after scattering"
input18="0 #SPECTRUM, use spectrum, ! was 1 initially!"
input19="0 #DIPOLE, add a dipole to the station, useful for st_type=0 and 2"
input20="0 #CONST_ATTENLENGTH, use constant attenuation length if ==1"
input21="$L #ATTEN_UP, this is the conjuction of the plot attenlength_up and attlength_down when setting REFLECT_RATE=0.5(3dB)"
input22="250 #ATTEN_DOWN, this is the average attenlength_down before Minna Bluff measurement(not used anymore except for CONST_ATTENLENGTH)"
input23="4 #NSIGMA, threshold of trigger"
input24="1 #ATTEN_FACTOR, change of the attenuation length"
input25="$Rval #REFLECT_RATE,power reflection rate at the ice bottom"
input26="0 #GZK, 1 means using GZK flux, 0 means E-2 flux"
input27="0 #FANFLUX, use fenfang's flux which only covers from 10^17 eV to 10^20 eV"
input28="0 #WIDESPECTRUM, use 10^16 eV to 10^21.5 eV as the energy spectrum, otherwise use 17-20"
input29="1 #SHADOWING"
input30="1 #DEPTH_DEPENDENT_N;0 means uniform firn, 1 means n_firn is a function of depth"
input31="0 #HEXAGONAL"
input32="1 #SIGNAL_FLUCT 1=add noise fluctuation to signal or 0=do not"
input33="4.0 #GAINV gain dependency"
input34="1 #TAUREGENERATION if 1=tau regeneration effect, if 0=original"
input35="3.0 #ST4_R radius in meters between center of station and antenna"
input36="350 #TNOISE noise temperature in Kelvin"
input37="80 #FREQ_LOW low frequency of LPDA Response MHz #was 100"
input38="1000 #FREQ_HIGH high frequency of LPDA Response MHz"
input39="/home/rajasekera.3/shelfmc/ShelfMC/temp/LP_gain_manual.txt #GAINFILENAME"
input40="$SD #STATION_DEPTH"
#######################################################################################################
cd $TMPDIR
mkdir $runName
cd $runName
initSeed=$seed
counter=0
for L in {500..1000..100} #attenuation length 500-1000
do
mkdir Atten_Up$L
cd Atten_Up$L
for R in {0..100..25} #Reflection Rate 0-1
do
mkdir ReflectionRate$R
cd ReflectionRate$R
if [ "$R" = "100" ]; then #fixing reflection rate value
Rval="1.0"
else
Rval="0.$R"
fi
for T in {500..2900..400} #Thickness of Ice 500-2900
do
mkdir IceThick$T
cd IceThick$T
for FT in {60..140..20} #Firn Thinckness 60-140
do
mkdir FirnThick$FT
cd FirnThick$FT
for SD in {0..200..50} #Station Depth
do
mkdir StationDepth$SD
cd StationDepth$SD
#####Do file operations###########################################
counter=$((counter+1))
echo "Counter = $counter ; L = $L ; R = $Rval ; T = $T ; FT = $FT ; SD = $SD " #print variables
#define changing lines
input21="$L #ATTEN_UP, this is the conjuction of the plot attenlength_up and attlength_down when setting REFLECT_RATE=0.5(3dB)"
input25="$Rval #REFLECT_RATE,power reflection rate at the ice bottom"
input10="$T #ICETHICK, thickness of ice including firn, 575m at Moore's Bay"
input13="$FT #FIRNDEPTH in meters"
input40="$SD #STATION_DEPTH"
for (( i=1; i<=$ppn;i++)) #make 20 setup files for 20 processors
do
mkdir Setup$i #make setup folder
cd Setup$i #go into setup folder
seed="$(($initSeed + $i -1))" #calculate seed for this iteration
input3="$seed #seed Seed for Rand3"
for j in {1..40} #print all input.txt lines
do
lineName=input$j
echo "${!lineName}" >> input.txt
done
cd ..
done
pwd=`pwd`
#create job file
echo '#!/bin/bash' >> run_shelfmc_multithread.sh
echo '#PBS -l nodes=1:ppn='$ppn >> run_shelfmc_multithread.sh
echo '#PBS -l walltime=00:05:00' >> run_shelfmc_multithread.sh #change walltime as necessary
echo '#PBS -N jude_'$runName'_job' >> run_shelfmc_multithread.sh #change job name as necessary
echo '#PBS -j oe' >> run_shelfmc_multithread.sh
echo '#PBS -A PCON0003' >> run_shelfmc_multithread.sh #change group if necessary
echo 'cd ' $tmpShelfmc >> run_shelfmc_multithread.sh
echo 'runName='$runName >> run_shelfmc_multithread.sh
for (( i=1; i<=$ppn;i++))
do
echo './shelfmc_stripped.exe $runName/'Atten_Up$L'/'ReflectionRate$R'/'IceThick$T'/'FirnThick$FT'/'StationDepth$SD'/Setup'$i' _'$i'$runName &' >> run_shelfmc_multithread.sh
done
# echo './shelfmc_stripped.exe $runName/'Atten_Up$L'/'ReflectionRate$R'/'IceThick$T'/'FirnThick$FT'/'StationDepth$SD'/Setup1 _01$runName &' >> run_shelfmc_multithread.sh
echo 'wait' >> run_shelfmc_multithread.sh
echo 'cd $runName/'Atten_Up$L'/'ReflectionRate$R'/'IceThick$T'/'FirnThick$FT'/'StationDepth$SD >> run_shelfmc_multithread.sh
echo 'for (( i=1; i<='$ppn';i++)) #20 iterations' >> run_shelfmc_multithread.sh
echo 'do' >> run_shelfmc_multithread.sh
echo ' cd Setup$i #cd into setup dir' >> run_shelfmc_multithread.sh
echo ' mv *.root ..' >> run_shelfmc_multithread.sh
echo ' cd ..' >> run_shelfmc_multithread.sh
echo 'done' >> run_shelfmc_multithread.sh
echo 'hadd Result_'$runName'.root *.root' >> run_shelfmc_multithread.sh
echo 'rm *ShelfMCTrees*' >> run_shelfmc_multithread.sh
chmod u+x run_shelfmc_multithread.sh # make executable
##################################################################
cd ..
done
cd ..
done
cd ..
done
cd ..
done
cd ..
done
cd
mv $WorkDir/$runName $HOME
|
| Attachment 3: scheduler.sh
|
#!/bin/bash
#Jude Rajasekera 3/20/17
tmpShelfmc=$HOME/shelfmc/ShelfMC #location of Shelfmc
runName=ParamSpaceScanDir #name of run
cd $tmpShelfmc #move to home directory
if [ ! -f ./jobList.txt ]; then #see if there is an existing job file
echo "Creating new job List"
for L in {500..1000..100} #attenuation length 500-1000
do
for R in {0..100..25} #Reflection Rate 0-1
do
for T in {500..2900..400} #Thickness of Ice 500-2900
do
for FT in {60..140..20} #Firn Thinckness 60-140
do
for SD in {0..200..50} #Station Depth
do
echo "cd $runName/Atten_Up$L/ReflectionRate$R/IceThick$T/FirnThick$FT/StationDepth$SD" >> jobList.txt
done
done
done
done
done
else
echo "Picking up from last job"
fi
numbLeft=$(wc -l < ./jobList.txt)
while [ $numbLeft -gt 0 ];
do
jobs=$(showq | grep "rajasekera.3") #change username here
echo '__________Current Running Jobs__________'
echo "$jobs"
echo ''
runningJobs=$(showq | grep "rajasekera.3" | wc -l) #change unsername here
echo Number of Running Jobs = $runningJobs
echo Number of jobs left = $numbLeft
if [ $runningJobs -le 6 ];then
line=$(head -n 1 jobList.txt)
$line
echo Submit Job && pwd
qsub run_shelfmc_multithread.sh
cd $tmpShelfmc
sed -i 1d jobList.txt
else
echo "Full Capacity"
fi
sleep 1
numbLeft=$(wc -l < ./jobList.txt)
done
|
|
|
24
|
Wed Jun 6 17:48:47 2018 |
Jorge Torres | How to build ROOT 6 on an OSC cluster | | Disclaimer: I wrote this for Owens, which I think will also work on Pitzer. I recommend following Steven's instructions, and use mine if it fails to build. J
1. Submit a batch job so the processing resources are not limited (change the project ID if needed.):
qsub -A PAS0654 -I -l nodes=1:ppn=4,walltime=2:00:00
2. Reset and load the following modules (copy and paste as it is):
module reset
module load cmake/3.7.2
module load python/2.7.latest
module load fftw3/3.3.5
3. Do echo $FFTW3_HOME and make sure it spits out "/usr/local/fftw3/intel/16.0/mvapich2/2.2/3.3.5". If it doesn't, do
export FFTW3_HOME=/usr/local/fftw3/intel/16.0/mvapich2/2.2/3.3.5
Otherwise, just do
export FFTW_DIR=$FFTW3_HOME
4. Do (Change DCMAKE_INSTALL_PREFIX and point it to the root source directory)
cmake -DCMAKE_C_COMPILER=`which gcc` \
-DCMAKE_CXX_COMPILER=`which g++` \
-DCMAKE_INSTALL_PREFIX=${HOME}/local/oakley/ROOT-6.12.06 \
-DBLAS_mkl_intel_LIBRARY=${MKLROOT}/lib/intel64 \
../root-6.12.06 2>&1 | tee cmake.log
It will configure root so it can be installed in the machine (takes about 5 minutes).
5. Once it is configured, do the following to build root (takes about 45 min)
make -j4 2>&1 | tee make.log
6. Once it's done, do
make install
In order to run it, now go into the directory, then cd bin. Once you're in there you should see a .sh called 'thisroot.sh'. Type 'source thisroot.sh'. You should now be able to type 'root' and it will run. Note that you must source this EVERY time you log into OSC. The smart thing to do would be to put this into your bash script.
(Second procedure from S. Prohira)
1. download ROOT: https://root.cern.ch/downloading-root (whatever the latest pro release is)
2. put the source tarball somewhere in your directory on ruby and expand it into the "source" folder
3. on ruby, open your ~/.bashrc file and add the following lines:
export CC="/usr/local/gnu/7.3.0/bin/gcc"
export CXX="/usr/local/gnu/7.3.0/bin/g++"
module load cmake
module load python
module load gnu/7.3.0
4. then run: source ~/.bashrc
5. make a "build" directory somewhere else on ruby called 'root' or 'root_build' and cd into that directory.
6. do: cmake /path/to/source/folder (e.g. the folder you expanded from the .tar file above. should finish with no errors.) here you can also include the -D flags that you want (such as minuit2 for the anita tools)
-for example, the ANITA tools need you to do: cmake -Dminuit2:bool=true /path/to/source/folder.
7. do: make -j4 (or the way that Jorge did it above, if you want to submit it as a batch job (and not be a jerk running a job on the login nodes like i did))
8. add the following line to your .bashrc file (or .profile, whatever startup file you prefer):
source /path/to/root/build/directory/bin/thisroot.sh
9. enjoy root!
|
|
|
28
|
Fri Oct 26 18:08:43 2018 |
Jorge Torres | Analyzing effective volumes | Analysis | Attaching some scripts that help processing the effective volumes. This is an extension of what Brian Clark did in a previous post (http://radiorm.physics.ohio-state.edu/elog/How-To/27)
There are 4 files attached:
- veff_aeff2.C and veff_aeff2.mk. veff_aeff2.C produces Veff_des$1.txt ($1 can be A or B or C). This file contains the following columns: energy, veff, veff_error, veff1 (PA), veff2 (LPDA), veff3 (bicone), respectively. However, the energies are not sorted.
-veff.sh: this bash executable runs veff_aeff2.C for all (that's what the "*" in the executable is for) the root output files, for a given design (A, B, C). You need to modify the location of your output files, though. Run like "./veff.sh A", which will execute veff_aeff2.C and produce the veff text files. Do the same for B or C.
-make_plot.py: takes Veff_des$1.txt, sorts energies out, plots the effective volumes vs. energies, and produces a csv file containing the veffs (just for the sake of copying and pastting on the spreadsheets). Run like "pyhton make_plot.py".
|
| Attachment 1: veff.sh
|
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_16.5.txt.run* &
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_17.txt.run* &
nohup ./veff_aeff2 3000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_17.5.txt.run* &
nohup ./veff_aeff2 5000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_18.txt.run* &
nohup ./veff_aeff2 5000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_18.5.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_19.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_19.5.txt.run* &
nohup ./veff_aeff2 7000 3000 $1 /users/PAS0654/osu8354/outputs_signal_noise/des"$1"/AraOut.des"$1"_20.txt.run* &
|
| Attachment 2: veff_aeff2.C
|
//////////////////////////////////////
//To calculate various veff and aeff
//Run as ./veff_aeff2 $RADIUS $DEPTH $FILE
//For example ./veff_aeff2 8000 3000 /data/user/ypan/bin/AraSim/trunk/outputs/AraOut.setup_single_station_2_energy_20.run0.root
//
/////////////////////////////////////
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TH1.h"
#include "TF1.h"
#include "TF2.h"
#include "TFile.h"
#include "TRandom.h"
#include "TRandom2.h"
#include "TRandom3.h"
#include "TTree.h"
#include "TLegend.h"
#include "TLine.h"
#include "TROOT.h"
#include "TPostScript.h"
#include "TCanvas.h"
#include "TH2F.h"
#include "TText.h"
#include "TProfile.h"
#include "TGraphErrors.h"
#include "TStyle.h"
#include "TMath.h"
#include <unistd.h>
#include "TVector3.h"
#include "TRotation.h"
#include "TSpline.h"
//#include "TObject.h"
#include "Tools.h"
#include "Constants.h"
#include "Vector.h"
#include "Position.h"
#include "EarthModel.h"
#include "IceModel.h"
#include "Efficiencies.h"
#include "Spectra.h"
#include "Event.h"
#include "Trigger.h"
#include "Detector.h"
#include "Settings.h"
#include "counting.hh"
#include "Primaries.h"
#include "signal.hh"
#include "secondaries.hh"
#include "Ray.h"
#include "RaySolver.h"
#include "Report.h"
using namespace std;
class EarthModel;
int main(int argc, char **argv){
//string readfile;
//readfile = string(argv[1]);
//readfile = "/data/user/ypan/bin/AraSim/branches/new_geom/outputs/AraOut20.0.root";
int ifile = 0;
double totweightsq;
double totweight;
int totnthrown;
int typetotnthrown[12];
double tottrigeff;
double sigma[12];
double typetotweight[12];
double typetotweightsq[12];
double totsigmaweight;
double totsigmaweightsq;
double volradius;
double voldepth;
const int nstrings = 9;
const int nantennas = 1;
double veff1 = 0.0;
double veff2 = 0.0;
double veff3 = 0.0;
double weight1 = 0.0;
double weight2 = 0.0;
double weight3 = 0.0;
double veffT[6], vefferrT[6], aeffT[6], aefferrT[6];
double veffF[3], vefferrF[3], aeffF[3], aefferrF[3];
double veffNu[2], vefferrNu[2], aeffNu[2], aefferrNu[2];
double veff, vefferr, aeff, aefferr, aeff2;
double pnu;
Detector *detector = 0;
//Settings *settings = 0;
//IceModel *icemodel = 0;
Event *event = 0;
Report *report = 0;
cout<<"construct detector"<<endl;
//TFile *AraFile=new TFile(readfile.c_str());
//TFile *AraFile=new TFile((outputdir+"/AraOut.root").c_str());
TChain *AraTree = new TChain("AraTree");
TChain *AraTree2 = new TChain("AraTree2");
TChain *eventTree = new TChain("eventTree");
//AraTree->SetBranchAddress("detector",&detector);
//AraTree->SetBranchAddress("settings",&settings);
//AraTree->SetBranchAddress("icemodel",&icemodel);
cout << "trees set" << endl;
for(ifile = 3; ifile < (argc - 1); ifile++){
AraTree->Add(string(argv[ifile + 1]).c_str());
AraTree2->Add(string(argv[ifile + 1]).c_str());
eventTree->Add(string(argv[ifile + 1]).c_str());
}
AraTree2->SetBranchAddress("event",&event);
AraTree2->SetBranchAddress("report",&report);
cout<<"branch detector"<<endl;
for(int i=0; i<12; i++) {
typetotweight[i] = 0.0;
typetotweightsq[i] = 0.0;
typetotnthrown[i] = 0;
}
totweightsq = 0.0;
totweight = 0.0;
totsigmaweight = 0.0;
totsigmaweightsq = 0.0;
totnthrown = AraTree2->GetEntries();
cout << "Total number of events: " << totnthrown << endl;
//totnthrown = settings->NNU;
//volradius = settings->POSNU_RADIUS;
volradius = atof(argv[1]);
voldepth = atof(argv[2]);
AraTree2->GetEntry(0);
pnu = event->pnu;
cout << "Energy " << pnu << endl;
for(int iEvt2=0; iEvt2<totnthrown; iEvt2++) {
AraTree2->GetEntry(iEvt2);
double sigm = event->Nu_Interaction[0].sigma;
int iflavor = (event->nuflavorint)-1;
int inu = event->nu_nubar;
int icurr = event->Nu_Interaction[0].currentint;
sigma[inu+2*icurr+4*iflavor] = sigm;
typetotnthrown[inu+2*icurr+4*iflavor]++;
if( (iEvt2 % 10000 ) == 0 ) cout << "*";
if(report->stations[0].Global_Pass<=0) continue;
double weight = event->Nu_Interaction[0].weight;
if(weight > 1.0){
cout << weight << "; " << iEvt2 << endl;
continue;
}
// cout << weight << endl;
totweightsq += pow(weight,2);
totweight += weight;
typetotweight[inu+2*icurr+4*iflavor] += weight;
typetotweightsq[inu+2*icurr+4*iflavor] += pow(weight,2);
totsigmaweight += weight*sigm;
totsigmaweightsq += pow(weight*sigm,2);
int trig1 = 0;
int trig2 = 0;
int trig3 = 0;
for (int i = 0; i < nstrings; i++){
if (i == 0 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig1++;
if (i > 0 && i < 5 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig2++;
if (i > 4 && i < 9 && report->stations[0].strings[i].antennas[0].Trig_Pass > 0) trig3++;
}
if ( trig1 > 0)//phase array
weight1 += event->Nu_Interaction[0].weight;
if ( trig2 > 1)//lpda
weight2 += event->Nu_Interaction[0].weight;
if (trig3 > 3)//bicone
weight3 += event->Nu_Interaction[0].weight;
}
tottrigeff = totweight / double(totnthrown);
double nnucleon = 5.54e29;
double vtot = PI * double(volradius) * double(volradius) * double(voldepth) / 1e9;
veff = vtot * totweight / double(totnthrown) * 4.0 * PI;
//vefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totweightsq)/totweight));
vefferr = sqrt(totweightsq) / totweight * veff;
aeff = vtot * (1e3) * nnucleon * totsigmaweight / double(totnthrown);
//aefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totsigmaweightsq)/totsigmaweight));
//aefferr = sqrt(SQ(sqrt(double(totnthrown))/double(totnthrown))+SQ(sqrt(totweightsq)/totweight));
aefferr = sqrt(totweightsq) / totweight * aeff;
double sigmaave = 0.0;
for(int iflavor=0; iflavor<3; iflavor++) {
double flavorweight = 0.0;
double flavorweightsq = 0.0;
double flavorsigmaave = 0.0;
int flavortotthrown = 0;
double temptotweightnu[2] = {0};
double tempsignu[2] = {0};
double temptotweight = 0.0;
for(int inu=0; inu<2; inu++) {
double tempsig = 0.0;
double tempweight = 0.0;
for(int icurr=0; icurr<2; icurr++) {
tempsig += sigma[inu+2*icurr+4*iflavor];
tempsignu[inu] += sigma[inu+2*icurr+4*iflavor];
tempweight += typetotweight[inu+2*icurr+4*iflavor];
flavorweight += typetotweight[inu+2*icurr+4*iflavor];
flavorweightsq += typetotweightsq[inu+2*icurr+4*iflavor];
temptotweight += typetotweight[inu+2*icurr+4*iflavor];
temptotweightnu[inu] += typetotweight[inu+2*icurr+4*iflavor];
flavortotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
//printf("Temp Sigma: "); cout << tempsig << "\n";
sigmaave += tempsig*(tempweight/totweight);
}
flavorsigmaave += tempsignu[0]*(temptotweightnu[0]/temptotweight)+tempsignu[1]*(temptotweightnu[1]/temptotweight);
veffF[iflavor] = vtot*flavorweight/double(flavortotthrown);
vefferrF[iflavor] = sqrt(flavorweightsq)/flavorweight;
//printf("Volume: %.9f*%.9f/%.9f \n",vtot,flavorweight,double(totnthrown));
aeffF[iflavor] = veffF[iflavor]*(1e3)*nnucleon*flavorsigmaave;
aefferrF[iflavor] = sqrt(flavorweightsq)/flavorweight;
}
for(int inu=0; inu<2; inu++) {
double tempsig = 0.0;
double tempweight = 0.0;
double tempweightsq = 0.0;
int nutotthrown = 0;
for(int iflavor=0; iflavor<3; iflavor++) {
for(int icurr=0; icurr<2; icurr++) {
tempweight += typetotweight[inu+2*icurr+4*iflavor];
tempweightsq += typetotweightsq[inu+2*icurr+4*iflavor];
nutotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
}
tempsig += sigma[inu+2*0+4*0];
tempsig += sigma[inu+2*1+4*0];
veffNu[inu] = vtot*tempweight/double(nutotthrown);
vefferrNu[inu] = sqrt(tempweightsq)/tempweight;
aeffNu[inu] = veffNu[inu]*(1e3)*nnucleon*tempsig;
aefferrNu[inu] = sqrt(tempweightsq)/tempweight;
}
double totalveff = 0.0;
double totalaeff = 0.0;
for(int inu=0; inu<2; inu++) {
for(int iflavor=0; iflavor<3; iflavor++) {
int typetotthrown = 0;
for(int icurr=0; icurr<2; icurr++) {
typetotthrown += typetotnthrown[inu+2*icurr+4*iflavor];
}
totalveff += veffT[iflavor+3*inu]*(double(typetotthrown)/double(totnthrown));
totalaeff += aeffT[iflavor+3*inu]*(double(typetotthrown)/double(totnthrown));
}
}
aeff2 = veff*(1e3)*nnucleon*sigmaave;
aeff = aeff2;
veff1 = weight1 / totnthrown * vtot * 4.0 * PI;
veff2 = weight2 / totnthrown * vtot * 4.0 * PI;
veff3 = weight3 / totnthrown * vtot * 4.0 * PI;
printf("\nvolthrown: %.6f; totweight: %.6f; Veff: %.6f +- %.6f\n", vtot, totweight, veff, vefferr);
printf("veff1: %.3f; veff2: %.3f; veff3: %.3f\n", veff1, veff2, veff3);
//string des = string(argv[4]);
char buf[100];
std::ostringstream stringStream;
stringStream << string(argv[3]);
std::string copyOfStr = stringStream.str();
snprintf(buf, sizeof(buf), "Veff_des%s.txt", copyOfStr.c_str());
FILE *fout = fopen(buf, "a+");
fprintf(fout, "%e, %.6f, %.6f, %.3f, %.3f, %.3f \n", pnu, veff, vefferr, veff1, veff2, veff3);
fclose(fout);
return 0;
}
|
| Attachment 3: veff_aeff2.mk
|
#############################################################################
## Makefile -- New Version of my Makefile that works on both linux
## and mac os x
## Ryan Nichol <rjn@hep.ucl.ac.uk>
##############################################################################
##############################################################################
##############################################################################
##
##This file was copied from M.readGeom and altered for my use 14 May 2014
##Khalida Hendricks.
##
##Modified by Brian Clark for use on CosTheta_NuTraject on 20 February 2015
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##
##############################################################################
##############################################################################
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o veff_aeff2.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc veff_aeff2.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = veff_aeff2
all : $(PROGRAMS)
veff_aeff2 : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
| Attachment 4: make_plot.py
|
# -*- coding: utf-8 -*-
import numpy as np
import sys
import matplotlib.pyplot as plt
from pylab import setp
from matplotlib.pyplot import rcParams
import csv
import pandas as pd
rcParams['mathtext.default'] = 'regular'
def read_file(finame):
fi = open(finame, 'r')
rdr = csv.reader(fi, delimiter=',', skipinitialspace=True)
table = []
for row in rdr:
# print(row)
energy = float(row[0])
veff = float(row[1])
veff_err = float(row[2])
veff1 = float(row[3])
veff2 = float(row[4])
veff3 = float(row[5])
row = {'energy':energy, 'veff':veff, 'veff_err':veff_err, 'veff1':veff1, 'veff2':veff2, 'veff3':veff3}
table.append(row)
df=pd.DataFrame(table)
df_ordered=df.sort_values('energy',ascending=True)
# print(df_ordered)
return df_ordered
def beautify_veff(this_ax):
sizer=20
xlow = 1.e16 #the lower x limit
xup = 2.e20 #the uppper x limit
ylow =1e-3 #the lower x limit
yup = 6.e1 #the uppper x limit
this_ax.set_xlabel('Energy [eV]',size=sizer) #give it a title
this_ax.set_ylabel('[V$\Omega]_{eff}$ [km$^3$sr]',size=sizer)
this_ax.set_yscale('log')
this_ax.set_xscale('log')
this_ax.tick_params(labelsize=sizer)
this_ax.set_xlim([xlow,xup]) #set the x limits of the plot
this_ax.set_ylim([ylow,yup]) #set the y limits of the plot
this_ax.grid()
this_legend = this_ax.legend(loc='upper left')
setp(this_legend.get_texts(), fontsize=17)
setp(this_legend.get_title(), fontsize=17)
def main():
"""
arasim_energies = np.array([3.16e+16, 1e+17, 3.16e+17, 1e+18, 3.16e+18, 1e+19, 3.16e+19, 1e+20])
arasim_energies2 = np.array([3.16e+16, 1e+17, 3.16e+17, 1e+18, 1e+19, 3.16e+19, 1e+20])
#arasim_desA_veff = np.array([0.080894,0.290695,0.943223,2.388708,4.070498,6.824112,10.506490,13.969418])
arasim_desA_veff_s = np.array([0.067384,0.289591,0.996509,2.464464,4.945600,8.735506,13.357300,18.751915])
# arasim_desA_veff_ice = np.array([0.066291,0.303620,0.927647,2.427554,4.962093,8.465895,13.425852,18.706528])
arasim_desA_error = np.array([0.008367,0.017401,0.032222,0.084223,0.119240,0.221266,0.272460,0.320456])
# arasim_desA_error_ice = np.array([0.008345,0.017748,0.031295,0.083747,0.119370,0.217717,0.273075,0.319872])
# arasim_desB_veff = np.array([0.124111,0.417102,1.310555,3.648494,4.070E+0,11.675189,18.393961,13.688909])
arasim_desB_veff_s = np.array([0.064937,0.355747,1.289947,3.821705,8.002805,15.352981,25.391282,18.009545])
# arasim_desB_veff_ice = np.array([0.080070,0.340927,1.369081,3.938550,8.211407,15.190858,25.066541,18.021033])
arasim_desB_error = np.array([0.008268,0.019317,0.036926,0.105445,0.152488,0.296617,0.377997,0.314314])
# arasim_desB_error_ice = np.array([0.009220,0.019144,0.037966,0.107189,0.154430,0.293728,0.375827,0.314244])
arasim_desA_veff1_sm = np.array([0.054,0.231,0.868,2.239,4.586,8.339,12.910,18.301])
arasim_desA_veff2_sm = np.array([0.009,0.036,0.098,0.323,0.705,1.167,2.169,3.130])
arasim_desA_veff3_sm = np.array([0.011,0.074,0.193,0.664,1.208,2.347,3.689,4.814])
arasim_desA_veff1 = np.array([0.053,0.282,1.108,3.462,7.486,14.613,24.682,17.557])
arasim_desA_veff2 = np.array([0.006,0.040,0.121,0.322,0.636,1.308,1.961,2.708])
arasim_desA_veff3 = np.array([0.006,0.065,0.188,0.638,1.187,2.270,3.301,4.322])
"""
veff_A=read_file("Veff_desA.txt")
veff_B=read_file("Veff_desB.txt")
print("desA is \n", veff_A)
print("desB is \n", veff_B)
dfA = veff_A[['veff','veff_err']]
dfB = veff_B[['veff','veff_err']]
dfA.to_csv('veffA.csv', sep='\t',index=False)
dfB.to_csv('veffB.csv', sep='\t',index=False)
fig = plt.figure(figsize=(11,8.5))
ax1 = fig.add_subplot(1,1,1)
ax1.plot(veff_A['energy'], veff_A['veff'],'bs-',label='Strawman',markersize=8,linewidth=2)
ax1.plot(veff_B['energy'], veff_B['veff'],'gs-',label='Punch @100 m',markersize=8,linewidth=2)
ax1.fill_between(veff_A['energy'], veff_A['veff']-veff_A['veff_err'], veff_A['veff']+veff_A['veff_err'], alpha=0.2, color='red')
ax1.fill_between(veff_B['energy'], veff_B['veff']-veff_B['veff_err'], veff_B['veff']+veff_B['veff_err'], alpha=0.2, color='red')
beautify_veff(ax1)
ax1.set_title("Punch vs Strawman, noise + signal",fontsize=20)
fig.savefig("desAB.png",edgecolor='none',bbox_inches="tight") #save the figure
fig2 = plt.figure(figsize=(11,8.5))
ax2 = fig2.add_subplot(1,1,1)
#Triggers plot
ax2.plot(veff_B['energy'], veff_B['veff1'],'g^-',label='Phased array (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff1'],'gs-',label='Phased array (Strawman)',markersize=8,linewidth=2)
ax2.plot(veff_B['energy'], veff_B['veff2'],'b^-',label='LPDAs (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff2'],'bs-',label='LPDAs (Strawman)',markersize=8,linewidth=2)
ax2.plot(veff_B['energy'], veff_B['veff3'],'y^-',label='Bicones (Punch)',markersize=8,linewidth=2)
ax2.plot(veff_A['energy'], veff_A['veff3'],'ys-',label='Bicones (Strawman)',markersize=8,linewidth=2)
ax2.set_title("Triggers contribution, noise + signal",fontsize=20)
beautify_veff(ax2)
fig2.savefig("desAB_triggers.png",edgecolor='none',bbox_inches="tight") #save the figure
main()
|
|
|
36
|
Mon Mar 4 14:11:07 2019 |
Jorge Torres | Submitting arrays of jobs on OSC | Analysis | PBS has the option of having arrays of jobs in case you want to easily submit multiple similar jobs. The only difference in them is the array index, which you can use in your PBS script to run each task with a different set of input arguments, or any other operation that requires a unique index.
You need to add the following lines to your submitter file:
#PBS -t array_min-array_max%increment
where "array_min/array_max" are integers that set lower and upper limit, respectively, of your "job loop" and increment lets you set the number of jobs that you want to submit simultaneously. For example:
submits an array with 100 jobs in it, but the system will only ever have 5 running at one time.
Here's an example of a script that submits to Pitzer a job array (from 2011-3000 in batches of 40) of a script named "make_fits_noise" that uses one core only. Make sure you use "#PBS -m n", otherwise you'll get tons of emails notyfing you about your jobs.
To delete the whole array, use "qdel JOB_ID[ ]". To delete one single instance, use "qdel JOB_ID[ARRAY_ID]".
More info: https://arc-ts.umich.edu/software/torque/job-arrays/
|
|
|
Draft
|
Fri Jan 31 10:43:52 2020 |
Jorge Torres | Mounting ARA software on OSC through CernVM File System | Software | OSC added ARA's CVMFS repository on Pitzer and Owens. This has been already done in UW's cluster thanks to Ben Hokanson-Fasig and Brian Clark. With CVMFS, all the dependencies are compiled and stored in a single folder (container), meaning that the user can just source the paths to the used environmental variables and not worry about installing them at all. This is very useful, since it usually takes a considerable amount of time to get those dependencies and properly install/debug the ARA software. To use the software, all you have to do is:
source /cvmfs/ara.opensciencegrid.org/trunk/centos7/setup.sh
To verify that the containter was correctly loaded, type
and see if the root display pops up. You can also go to /cvmfs/ara.opensciencegrid.org/trunk/centos7/source/AraSim and execute ./AraSim
Because of it being a container, the permissions are read-only. This means that if you want to do any modifications to the existing code, you'll have to copy the piece of code that you want, and change the enviromental variables of that package, in this case $ARA_UTIL_INSTALL_DIR, which is the destination where you want your executables, libraries and such, installed.
Libraries and executables are stored here, in case you want to reference those dependencies as your environmental variables: /cvmfs/ara.opensciencegrid.org/trunk/centos7/
Even if you're not in the ARA collaboration, you can benefit from this through the fact that ROOT6 is installed and compiled in the container. In order to use it you just need to run the same bash command, and ROOT 6 will be available for you to use.
Feel free to email any questions to Brian Clark or myself.
--------
Technical notes:
The ARA software was compiled with gcc version 4.8.5. In OSC, that compiler can be loaded by doing module load gnu/4.8.5. If you're using any other compiler, you'll get a warning telling you that if you do any compilation against the ARA software, you may need to add the -D_GLIBCXX_USE_CXX11_ABI=0 flag to your Make file. |
|
|
49
|
Thu Sep 14 22:30:06 2023 |
Jason Yao | How to profile a C++ program | Software | This guide is modified from section (d) of the worksheet inside Module 10 of Phys 6810 Computational Physics (Spring 2023).
NOTE: gprof does not work on macOS. Please use a linux machine (such as OSC)
To use gprof, compile and link the relevant codes with the -pg option:
Take a look at the Makefile make_hello_world and modify both the CFLAGS and LDFLAGS lines to include -pg
Compile and link the script by typing
make -f make_hello_world
Execute the program
./hello_world.x
With the -pg flags, the execution will generate a file called gmon.out that is used by gprof.
The program has to exit normally (e.g. we can't stop with a ctrl-C).
Warning: Any existing gmon.out file will be overwritten.
Run gprof and save the output to a file (e.g., gprof.out) by
gprof hello_world.x > gprof.out
We should at this point see a text file called gprof.out which contains the profile of hello_world.cpp
vim gprof.out |
| Attachment 1: hello_world.cpp
|
#include <iostream>
#include <thread>
#include <chrono>
#include <cmath>
using namespace std;
void nap(){
// usleep(3000);
// this_thread::sleep_for(30000ms);
for (int i=0;i<1000000000;i++){
double j = sqrt(i^2);
}
}
int main(){
cout << "taking a nap" << endl;
nap();
cout << "hello world" << endl;
}
|
| Attachment 2: make_hello_world
|
SHELL=/bin/sh
# Note: Comments start with #. $(FOOBAR) means: evaluate the variable
# defined by FOOBAR= (something).
# This file contains a set of rules used by the "make" command.
# This makefile $(MAKEFILE) tells "make" how the executable $(COMMAND)
# should be create from the source files $(SRCS) and the header files
# $(HDRS) via the object files $(OBJS); type the command:
# "make -f make_program"
# where make_program should be replaced by the name of the makefile.
#
# Programmer: Dick Furnstahl (furnstahl.1@osu.edu)
# Latest revision: 12-Jan-2016
#
# Notes:
# * If you are ok with the default options for compiling and linking, you
# only need to change the entries in section 1.
#
# * Defining BASE determines the name for the makefile (prepend "make_"),
# executable (append ".x"), zip archive (append ".zip") and gzipped
# tar file (append ".tar.gz").
#
# * To remove the executable and object files, type the command:
# "make -f $(MAKEFILE) clean"
#
# * To create a zip archive with name $(BASE).zip containing this
# makefile and the SRCS and HDRS files, type the command:
# "make -f $(MAKEFILE) zip"
#
# * To create a gzipped tar file with name $(BASE).tar.gz containing this
# makefile and the source and header files, type the command:
# "make -f $(MAKEFILE) tarz"
#
# * Continuation lines are indicated by \ with no space after it.
# If you get a "missing separator" error, it is probably because there
# is a space after a \ somewhere.
#
###########################################################################
# 1. Specify base name, source files, header files, input files
###########################################################################
# The base for the names of the makefile, executable command, etc.
BASE= hello_world
# Put all C++ (or other) source files here. NO SPACES after continuation \'s.
SRCS= \
hello_world.cpp
# Put all header files here. NO SPACES after continuation \'s.
HDRS= \
# Put any input files you want to be saved in tarballs (e.g., sample files).
INPFILE= \
###########################################################################
# 2. Generate names for object files, makefile, command to execute, tar file
###########################################################################
# *** YOU should not edit these lines unless to change naming conventions ***
OBJS= $(addsuffix .o, $(basename $(SRCS)))
MAKEFILE= make_$(BASE)
COMMAND= $(BASE).x
TARFILE= $(BASE).tar.gz
ZIPFILE= $(BASE).zip
###########################################################################
# 3. Commands and options for different compilers
###########################################################################
#
# Compiler parameters
#
# CXX Name of the C++ compiler to use
# CFLAGS Flags to the C++ compiler
# CWARNS Warning options for C++ compiler
# F90 Name of the fortran compiler to use (if relevant)
# FFLAGS Flags to the fortran compiler
# LDFLAGS Flags to the loader
# LIBS A list of libraries
#
CXX= g++
CFLAGS= -g
CWARNS= -Wall -W -Wshadow -fno-common
MOREFLAGS= -Wpedantic -Wpointer-arith -Wcast-qual -Wcast-align \
-Wwrite-strings -fshort-enums
# add relevant libraries and link options
LIBS=
# LDFLAGS= -lgsl -lgslcblas
LDFLAGS=
###########################################################################
# 4. Instructions to compile and link, with dependencies
###########################################################################
all: $(COMMAND)
.SUFFIXES:
.SUFFIXES: .o .mod .f90 .f .cpp
#%.o: %.mod
# This is the command to link all of the object files together.
# For fortran, replace CXX by F90.
$(COMMAND): $(OBJS) $(MAKEFILE)
$(CXX) -o $(COMMAND) $(OBJS) $(LDFLAGS) $(LIBS)
# Command to make object (.o) files from C++ source files (assumed to be .cpp).
# Add $(MOREFLAGS) if you want additional warning options.
%.o: %.cpp $(HDRS) $(MAKEFILE)
$(CXX) -c $(CFLAGS) $(CWARNS) -o $@ $<
# Commands to make object (.o) files from Fortran-90 (or beyond) and
# Fortran-77 source files (.f90 and .f, respectively).
.f90.mod:
$(F90) -c $(F90FLAGS) -o $@ $<
.f90.o:
$(F90) -c $(F90FLAGS) -o $@ $<
.f.o:
$(F90) -c $(FFLAGS) -o $@ $<
##########################################################################
# 5. Additional tasks
##########################################################################
# Delete the program and the object files (and any module files)
clean:
/bin/rm -f $(COMMAND) $(OBJS)
/bin/rm -f $(MODIR)/*.mod
# Pack up the code in a compressed gnu tar file
tarz:
tar cfvz $(TARFILE) $(MAKEFILE) $(SRCS) $(HDRS) $(MODIR) $(INPFILE)
# Pack up the code in a zip archive
zip:
zip -r $(ZIPFILE) $(MAKEFILE) $(SRCS) $(HDRS) $(MODIR) $(INPFILE)
##########################################################################
# That's all, folks!
##########################################################################
|
|
|
50
|
Wed Jun 12 12:10:05 2024 |
Jacob Weiler | How to install AraSim on OSC | Software | # Installing AraSim on OSC
Readding this because I realized it was deleted when I went looking for it 
Quick Links:
- https://github.com/ara-software/AraSim # AraSim github repo (bottom has installation instructions that are sort of right)
- Once AraSim is downloaded: AraSim/UserGuideTex/AraSimGuide.pdf (manual for AraSim) might have to be downloaded if you can't view pdf's where you write code
Step 1:
We need to add in the dependancies. AraSim needs multiple different packages to be able to run correctly. The easiest way on OSC to get these without a headache is to add the following to you .bashrc for your user.
cvmfs () {
module load gnu/4.8.5
export CC=`which gcc`
export CXX=`which g++`
if [ $# -eq 0 ]; then
local version="trunk"
elif [ $# -eq 1 ]; then
local version=$1
else
echo "cvmfs: takes up to 1 argument, the version to use"
return 1
fi
echo "Loading cvmfs for AraSim"
echo "Using /cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
source "/cvmfs/ara.opensciencegrid.org/${version}/centos7/setup.sh"
#export JUPYTER_CONFIG_DIR=$HOME/.jupyter
#export JUPYTER_PATH=$HOME/.local/share/jupyter
#export PYTHONPATH=/users/PAS0654/alansalgo1/.local/bin:/users/PAS0654/alansalgo1/.local/bin/pyrex:$PYTHONPATH
}
If you want to view my bashrc
- /users/PAS1977/jacobweiler/.bashrc
Reload .bashrc
- source ~/.bashrc
Step 2:
Go to directory that you want to put AraSim and type:
- git clone https://github.com/ara-software/AraSim.git
This will download the github repo
Step 3:
We need to use make and load sourcing
- cd AraSim
- cvmfs
- make
wait and it should compile the code
Step 4:
We want to do a test run with 100 neutrinos to make sure that it does *actually* run
Try: - ./AraSim SETUP/setup.txt
This errored for me (probably you as well)
Switch from frequency domain to time domain in the setup.txt
- cd SETUP
- open setup.txt
- scroll to bottom
- Change SIMULATION_MODE = 1
- save
- cd ..
- ./AraSim SETUP/setup.txt
This should run quickly and now you have AraSim setup! |
|
|
40
|
Thu Jul 25 16:50:43 2019 |
Dustin Nguyen | Advice (not mine) on writing HEP stuff | Other | PDF of advice by Andy Buckley (U Glasgow) on writing a HEP thesis (and presumably HEP papers too) that was forwarded by John Beacom to the CCAPP mailing list a few months back. |
| Attachment 1: thesis-writing-gotchas.pdf
|
|
|
17
|
Mon Nov 20 08:31:48 2017 |
Brian Clark and Oindree Banerjee | Fit a Function in ROOT | Analysis | Sometimes you need to fit a function to a histogram in ROOT. Attached is code for how to do that in the simple case of a power law fit.
To run the example, you should type "root fitSnr.C" in the command line. The code will access the source histogram (hstrip1nsr.root, which is actually ANITA-2 satellite data). The result is stripe1snrfit.png. |
| Attachment 1: fitSnr.C
|
#include "TF1.h"
void fitSnr();
void fitSnr()
{
gStyle->SetLineWidth(4); //set some style parameters
TFile *stripe1file = new TFile("hstripe1snr.root"); //import the file containing the histogram to be fit
TH1D *hstripe1 = (TH1D*)stripe1file->Get("stripe1snr"); //get the histogram to be fit
TCanvas c1("c1","c1",1000,800); //make a canvas
hstripe1->Draw(""); //draw it
c1.SetLogy(); //set a log axis
//need to declare an equation
//I want to fit for two paramters, in the equation these are [0] and [1]
//so, you'll need to re-write the equation to whatever you're trying to fit for
//but ROOT wants the variables to find to be given as [0], [1], [2], etc.
char equation1[150]; //declare a container for the equation
sprintf(equation1,"([0]*(x^[1]))"); //declare the equation
TF1 *fit1 = new TF1("PowerFit",equation1,20,50); //create a function to fit with, with the range being 20 to 50
//now, we need to set the initial parameters of the fit
//fit->SetParameter(0,H->GetRMS()); //this should be a good starting place for a standard deviation like variable
//fit->SetParameter(1,H->GetMaximum()); //this should be a good starting place for amplitude like variable
fit1->SetParameter(0,60000.); //for our example, we will manually choose this
fit1->SetParameter(1,-3.);
hstripe1->Fit("PowerFit","R"); //actually do the fit;
fit1->Draw("same"); //draw the fit
//now, we want to print out some parameters to see how good the fit was
cout << "par0 " << fit1->GetParameter(0) << " par1 " << fit1->GetParameter(1) << endl;
cout<<"chisquare "<<fit1->GetChisquare()<<endl;
cout<<"Ndf "<<fit1->GetNDF()<<endl;
cout<<"reduced chisquare "<<double(fit1->GetChisquare())/double(fit1->GetNDF())<<endl;
cout<<" "<<endl;
c1.SaveAs("stripe1snrfit.png");
}
|
| Attachment 2: hstripe1snr.root
|
| Attachment 3: stripe1snrfit.png
| 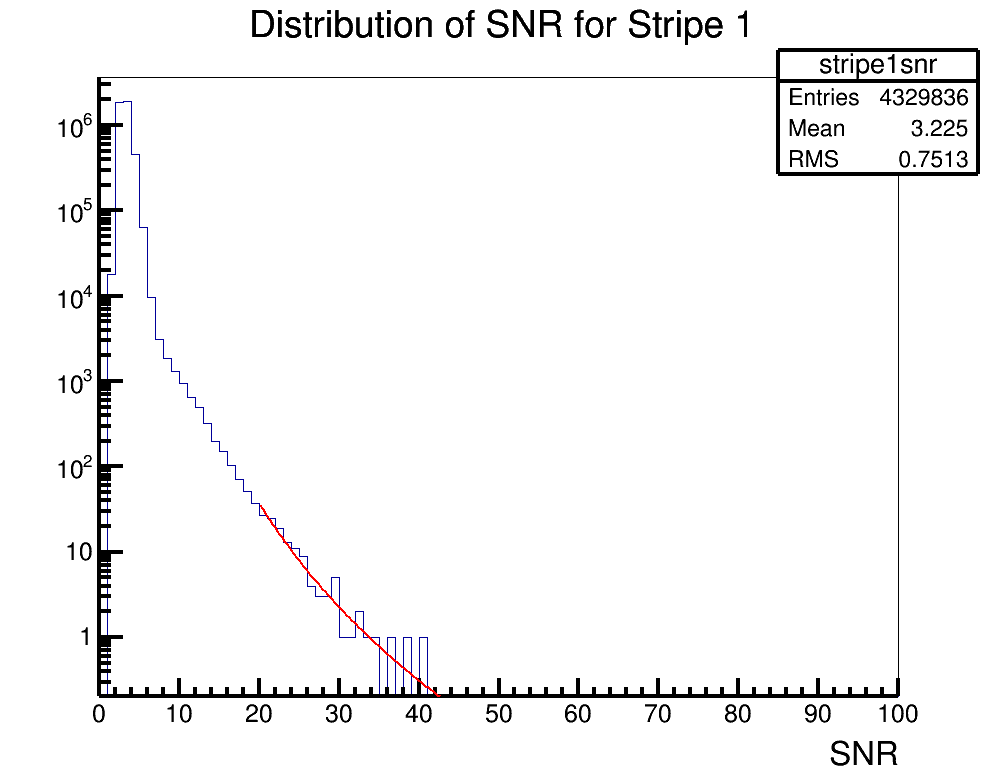 |
|
|
23
|
Wed Jun 6 08:54:44 2018 |
Brian Clark and Oindree Banerjee | How to Access Jacob's ROOT6 on Oakley | | Source the attached env.sh file. Good to go! |
| Attachment 1: env.sh
|
export ROOTSYS=/users/PAS0174/osu8620/root-6.08.06
eval 'source /users/PAS0174/osu8620/root-6.08.06/builddir/bin/thisroot.sh'
export LD_INCLUDE_PATH=/users/PAS0174/osu8620/cint/libcint/build/include:$LD_INCLUDE_PATH
module load fftw3/3.3.5
module load gnu/6.3.0
module load python/3.4.2
module load cmake/3.7.2
#might need this, but probably not
#export CC=/usr/local/gcc/6.3.0/bin/gcc
|
|
|
Draft
|
Fri Jul 28 17:57:06 2017 |
Brian Clark and Ian Best | | | |
|
|
3
|
Wed Mar 22 18:01:23 2017 |
Brian Clark | Advice for Using the Ray Trace Correlator | Analysis | If you are trying to use the Ray Trace Correlator with AraRoot, you will probably encounter some issues as you go. Here is some advice that Carl Pfendner found, and Brian Clark compiled.
Please note that it is extremely important that your AntennaInfo.sqlite table in araROOT contain the ICRR versions of both Testbed and Station1. Testbed seems to have fallen out of practice of being included in the SQL table. Also, Station1 is the ICRR (earliest) version of A1, unlike the ATRI version which is logged as ARA01. This will cause seg faults in the intial setup of the timing and geometry arrays that seem unrelated to pure geometry files. If you get a seg-fault in the "setupSizes" function or the Detector call of the "setupPairs" function, checking your SQL file is a good idea. araROOT branch 3.13 has such a source table with Testbed and Station1 included.
Which combination of Makefile/Makefile.arch/StandardDefinitions.mk works can be machine specific (frustratingly). Sometimes the best StandardDefinitions.mk is found int he make_timing_arrays example.
Common Things to Check
1: Did you "make install" the Ray Trace Correlator after you made it?
2: Do you have the setup.txt file?
3: Do you have the "data" directory?
Common Errors
1: If the Ray Trace Correlator compiles, and you execute a binary, and get the following:
******** Begin Correlator ********, this one!
Pre-icemodel test
terminate called after throwing an instance of 'std::out_of_range'
what(): basic_string::substr
Aborted
Check to make sure have the "data" directory.
|
|
|
16
|
Thu Oct 26 08:44:58 2017 |
Brian Clark | Find Other Users on OSC | Other | Okay, so our group has two "project" spaces on OSC (the Ohio Supercomputer). The first is for Amy's group, and is a project workspace called "PAS0654". The second is the CCAPP Condo (literally, CCAPP has some pre-specified rental time, and hence "condo") on OSC, and this is project PCON0003.
When you are signed up for the supercomputer, one of two things happen:
- You will be given a username under the PAS0654 group, and in which case, your username will be something like osu****. Connolly home drive is /users/PAS0654/osu****. Beatty home drive is /users/PAS0174/osu****. CCAPP home drive is /users/PCON0003/pcon****.
- You will be given a username under the PCON0003 group, and in which case, your username will be something like cond****.
If you are given a osu**** username, you must make sure to be added to the CCAPP Condo so that you can use Ruby compute resources. It will not be automatic.
Some current group members, and their OSC usernames. In parenthesis is the project space they are found in.
Current Users
osu0673: Brian Clark (PAS0654)
cond0068: Jorge Torres-Espinosa (PCON0003)
osu8619: Keith McBride (PAS0174)
osu9348: Julie Rolla (PAS0654)
osu9979: Lauren Ennesser (PAS0654)
osu6665: Amy Connolly (PAS0654)
Past Users
osu0426: Oindree Banerjee (PAS0654)
osu0668: Brian Dailey (PAS0654)
osu8620: Jacob Gordon (PAS0174)
osu8386: Sam Stafford (ANITA analysis in /fs/scratch/osu8386) (PAS0174)
cond0091: Judge Rajasekera (PCON0003) |
|
|
18
|
Tue Dec 12 17:38:36 2017 |
Brian Clark | Data Analysis in R from day w/ Brian Connolly | Analysis | On Nov 28 2017, Brian Connolly came and visited and taught us how to do basic data analysis in R.
He in particular showed us how to do a Linear Discriminant Analysis (LDA) and Principal Component Analysis (PCA).
Attached are three of the data files Carl Pfendner prepared for us to analyze (ARA data, including simulation, force triggers, and RF triggers).
Also attached is some R code that shows how to set up the LDA and the PCA and how to plot their result. You are meant to run each line of the code in r by hand (this is not a functioning R script I don't think).
Go here (https://www.r-project.org/) to learn how to download R. You will also probably have to download GGFortify. To do that, open an r session, and type "install.packages('ggfortify')". |
| Attachment 1: pca_and_lda.R
|
Notes
#First we need to read in the data
dataSim <- read.table("output.txt.Simulation",sep=" ",header=TRUE)
dataRF <- read.table("output.txt.RFTriggers",sep=" ",header=TRUE)
dataPulser <- read.table("output.txt.CalPulsers",sep=" ",header=TRUE)
#Now we combine them into one data object
data <- rbind(dataRF,dataSim)
#Now we actually have to specify that we
#want to use the first 10 columns
data <- data[,1:10]
#make a principal component analysis object
pca <- prcomp(data,center=TRUE,scale=TRUE,retx=TRUE)
#Load a plotting library
library(ggfortify)
#then can plot
autplot(prcomp(data))
#or, we can plot like this
#to get some color data points
labels<-c(rep(0,nrow(dataSim)),rep(1,nrow(dataRF)))
plot(pca$x[,1],pca$x[,2],col=labels+1)
#we can also do an LDA analysis
#we need to import the MASS library
library(MASS)
#and now we make the lda object
lda(data,grouping=labels)
#didn't write down any plotting unfortunately.
|
| Attachment 2: data.zip
|
|
|
19
|
Mon Mar 19 12:27:59 2018 |
Brian Clark | How To Do an ARA Monitoring Report | Other | So, ARA has five stations down in the ice that are taking data. Weekly, a member of the collaboration checks on the detectors to make sure that they are healthy.
This means things like making sure they are triggering at approximately the right rates, are taking cal pulsers, that the box isn't too hot, etc.
Here are some resources to get you started. Usual ara username and password apply in all cases.
Also, the page where all of the plots live is here: http://aware.wipac.wisc.edu/
Thanks, and good luck monitoring! Ask someone whose done it before when in doubt.
Brian |
|
|
20
|
Tue Mar 20 09:24:37 2018 |
Brian Clark | Get Started with Making Plots for IceMC | Software | |
Second, here is the page for the software IceMC, which is the Monte Carlo software for simulating neutrinos for ANITA.
On that page are good instructions for downloading the software and how to run it. You will have the choice of running it on a (1) a personal machine (if you want to use your personal mac or linux machine), (2) a queenbee laptop in the lab, or (3) on a kingbee account which I will send an email about shortly. Running IceMC will require a piece of statistics software called ROOT that can be somewhat challenging to install--it is already installed on Kingbee and OSC, so it is easier to get started there. If you want to use Kingbee, just try downloading and running. If you want to use OSC, you're first going to need to follow instructions to access a version installed on OSC. Still getting that together.
So, familiarize yourself with the command line, and then see if you can get ROOT and IceMC installed and running. Then plots. |
|
|
21
|
Fri Mar 30 12:06:11 2018 |
Brian Clark | Get icemc running on Kingbee and Unity | Software | So, icemc has some needs (like Mathmore) and preferably root 6 that aren't installed on kingbeen and unity.
Here's what I did to get icecmc running on kingbee.
Throughout, $HOME=/home/clark.2668
- Try to install new version fo ROOT (6.08.06, which is the version Jacob uses on OSC) with CMAKE. Failed because Kingbee version of cmake is too old.
- Downloaded new version of CMAKE (3.11.0), failed because kingbee doesn't have C++11 support.
- Downloaded new version of gcc (7.33) and installed that in $HOME/QCtools/source/gcc-7.3. So I installed it "in place".
- Then, compiled the new version of CMAKE, also in place, so it's in $HOME/QCtools/source/cmake-3.11.0.
- Then, tried to compile ROOT, but it got upset because it couldn't find CXX11; so I added "export CC=$HOME/QCtools/source/gcc-7.3/bin/gcc" and then it could find it.
- Then, tried to compile ROOT, but couldn't because ROOT needs >python 2.7, and kingbee has python 2.6.
- So, downloaded latest bleeding edge version of python 3 (pyton 3.6.5), and installed that with optimiation flags. It's installed in $HOME/QCtools/tools/python-3.6.5-build.
- Tried to compile ROOT, and realized that I need to also compile the shared library files for python. So went back and compiled with --enable-shared as an argument to ./configure.
- Had to set the python binary, include, and library files custom in the CMakeCache.txt file.
|
|
|
22
|
Sun Apr 29 21:44:15 2018 |
Brian Clark | Access Deep ARA Station Data | Analysis | Quick C++ program for pulling waveforms out of deep ARA station data. If you are using AraRoot, you would put this inside your "analysis" directory and add it to your CMakeLists.txt. |
| Attachment 1: plot_deep_station_event.cxx
|
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
//// plot_event.cxx
//// plot deep station event
////
//// Apr 2018, clark.2668@osu.edu
////////////////////////////////////////////////////////////////////////////////
//Includes
#include <iostream>
//AraRoot Includes
#include "RawAtriStationEvent.h"
#include "UsefulAtriStationEvent.h"
#include "AraEventCalibrator.h"
//ROOT Includes
#include "TTree.h"
#include "TFile.h"
#include "TGraph.h"
#include "TCanvas.h"
using namespace std;
int main(int argc, char **argv)
{
//check to make sure they've given me a run and a pedestal
if(argc<2) {
std::cout << "Usage\n" << argv[0] << " <run file>\n";
std::cout << "e.g.\n" << argv[0] << " event1841.root\n";
return 0;
}
char pedFileName[200];
sprintf(pedFileName, "%s", argv[1]);
printf("------------------------------------------------------------------------\n");
printf("%s\n", argv[0]);
printf("runFileName %s\n", argv[1]);
printf("------------------------------------------------------------------------\n");
TFile *fp = TFile::Open(argv[2]);
if(!fp) {
std::cerr << "Can't open file\n";
return -1;
}
TTree *eventTree = (TTree*) fp->Get("eventTree");
if(!eventTree) {
std::cerr << "Can't find eventTree\n";
return -1;
}
RawAtriStationEvent *rawAtriEvPtr = 0; //empty pointer
eventTree->SetBranchAddress("event",&rawAtriEvPtr); //set the branch address
Int_t run_num; //run number of event
eventTree->SetBranchAddress("run", &run_num); //set the branch address
int numEntries=eventTree->GetEntries(); //get the number of events
int stationId=0;
eventTree->GetEntry(0);
stationId = rawAtriEvPtr->stationId; //assign the statio id number
AraEventCalibrator *calib = AraEventCalibrator::Instance(); //make a calibrator
for(int event=0;event<numEntries;event++) {
//for(int event=0;event<700;event++) {
eventTree->GetEntry(event); //get the event
int evt_num = rawAtriEvPtr->eventNumber; //check the event umber
if(rawAtriEvPtr->isCalpulserEvent()==0) continue; //bounce out if it's not a cal pulser
UsefulAtriStationEvent *realAtriEvPtr_fullcalib = new UsefulAtriStationEvent(rawAtriEvPtr, AraCalType::kLatestCalib); //make the event
TGraph *waveforms[16]={0};
for(int i=0; i<16; i++){
waveforms[i]=realAtriEvPtr_fullcalib->getGraphFromRFChan(i);
}
TCanvas *canvas = new TCanvas("","",1000,1000);
canvas->Divide(4,4);
for(int i=0; i<16; i++){
canvas->cd(i+1);
waveforms[i]->Draw("alp");
}
char title[200];
sprintf(title,"waveforms_run%d_event%d.pdf",stationId,run_num,evt_num);
canvas->SaveAs(title);
delete canvas;
}
}
|
|
|
26
|
Sun Aug 26 19:23:57 2018 |
Brian Clark | Get a quick start with AraSim on OSC Oakley | Software | These are instructions I wrote for Rishabh Khandelwal to facilitate a "fast" start on Oakley at OSC. It was to help him run AraSim in batch jobs on Oakley.
It basically has you use software dependencies that I pre-installed on my OSC account at /users/PAS0654/osu0673/PhasedArraySimulation.
It also gives a "batch_processing" folder with examples for how to successfully run AraSim batch jobs (with correct output file management) on Oakley.
Sourcing these exact dependencies will not work on Owens or Ruby, sorry. |
| Attachment 1: forRishabh.tar.gz
|
|
|
27
|
Mon Oct 1 19:06:59 2018 |
Brian Clark | Code to Compute Effective Volumes in AraSim | Analysis | Here is some C++ code and an associated makefile to find effective volumes from AraSim output files.
It computes error bars on the effective volumes using the relevant AraSim function.
Compile like "make -f veff.mk"
Run like "./veff thrown_radius thrown_depth AraOut.1.root AraOut.2.root...." |
| Attachment 1: veff.cc
|
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include <cstdio>
#include <cstdlib>
#include <unistd.h>
#include <cstring>
#include <unistd.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TFile.h"
#include "TTree.h"
#include "TMath.h"
#include "Event.h"
#include "Detector.h"
#include "Report.h"
#include "Vector.h"
#include "counting.hh"
using namespace std;
//Get effective volumes with error bars
class EarthModel;
int main(int argc, char **argv)
{
gStyle->SetOptStat(111111);
gStyle->SetOptDate(1);
if(argc<4){
cout<<"Not enough arguments! Abort run."<<endl;
cout<<"Run like: ./veff volradius voldepth AraOut1.root AraOut2.rot ..."<<endl;
return -1;
}
double volradius = atof(argv[1]);
double voldepth = atof(argv[2]);
TChain *AraTree = new TChain("AraTree");
TChain *AraTree2 = new TChain("AraTree2");
for(int i=3; i<argc;i++){
AraTree->Add(string(argv[i]).c_str());
AraTree2->Add(string(argv[i]).c_str());
}
Report *report = 0;
Event *event=0;
AraTree2->SetBranchAddress("report",&report);
AraTree2->SetBranchAddress("event",&event);
int totnthrown = AraTree2->GetEntries();
cout << "Total number of events: " << totnthrown << endl;
int NBINS=10;
double eventsfound_binned[NBINS];
for(int i=0; i<NBINS; i++) eventsfound_binned[i]=0.;
double totweight=0;
for(int iEvt2=0; iEvt2<totnthrown; iEvt2++){
AraTree2->GetEntry(iEvt2);
if(report->stations[0].Global_Pass<=0) continue;
double weight = event->Nu_Interaction[0].weight;
if(weight > 1.0) continue;
totweight += weight;
int index_weights = Counting::findWeightBin(log10(weight));
if(index_weights<NBINS) eventsfound_binned[index_weights]++;
}
double error_minus=0.;
double error_plus=0.;
Counting::findErrorOnSumWeights(eventsfound_binned,error_plus,error_minus);
double vtot = TMath::Pi() * double(volradius) * double(volradius) * double(voldepth) / 1.e9; //answer in km^3
double veff = vtot * totweight / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
double veff_p = vtot * (error_plus) / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
double veff_m = vtot * (error_minus) / double(totnthrown) * 4. * TMath::Pi(); //answer in km^3 sr
printf("volthrown: %.6f \n totweight: %.6f + %.6f - %.6f \n Veff: %.6f + %.6f - %.6f \n",
vtot,
totweight, error_plus, error_minus,
veff, veff_p, veff_m
);
return 0;
}
|
| Attachment 2: veff.mk
|
#############################################################################
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o veff.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc veff.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = veff
all : $(PROGRAMS)
veff : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
|
|
29
|
Fri Nov 9 00:44:09 2018 |
Brian Clark | Transfer files from IceCube Data Warehouse to OSC | | Brian had to move ~7 TB of data from the IceCube data warehouse to OSC.
To do this, he used the gridftp software. The advantage is that griftp is optimized for large file transfers, and will manage data transfer better than something like scp or rsync.
Note that this utilizes the gridftp software installed on OSC, but doesn't formally use the globus end point described here: https://www.osc.edu/resources/getting_started/howto/howto_transfer_files_using_globus_connect. This is because IceCube doesn't have a formal globus endpoint to connect too. The formal globus endpoint would have been even easier if it was available, but oh well...
Setup goes as follows:
- Follow the IceCube instructions for getting an OSG certificate
- Go through CILogon (https://cilogon.org/) and generate and download a certificate
- Install your certificate on the IceCube machines
- Move your certificate to the following place on IceCube:
./globus/usercred.p12
- Change the permissions on this certificate:
chmod 600 ./globus/usercred.p12
- Get your "subject" to this key:
openssl pkcs12 -in .globus/usercred.p12 -nokeys | grep subject
- Copy the subject line into your IceCube LDAP account
- Select "Edit your profile"
- Enter IceCube credentials
- Paste the subject into the "x509 Subject DN" box
- Install your certificates on the OSC machines
- Follow the same instructions as for IceCube to install the globus credentials, but you don't need to do the IceCube LDAP part
How to actually make a transfer:
- Initialize a proxy certificate on OSC: grid-proxy-init -bits 1024
- Use globus-url-copy to move a file, for example:
globus-url-copy -r gsiftp://gridftp.icecube.wisc.edu/data/wipac/ARA/2016/unblinded/L1/ARA03/ 2016/ &
- I'm using the command "globus-url-copy"
- "-r" says to transfer recursively
- "gsiftp://gridftp.icecube.wisc.edu/data/wipac/ARA/2016/ublinded/L1/ARA03/" is the entire directory I'm trying to copy
- "2016/" is the directory I'm copying them to
- "&" says do this in the background once launched
- Note that it's kind of syntatically picky:
- To copy a directory, the source path name must end in "/"
- To copy a directory, the destination path name must also end in "/"
|
|
|
30
|
Tue Nov 27 09:43:37 2018 |
Brian Clark | Plot Two TH2Ds with Different Color Palettes | Analysis | Say you want to plot two TH2Ds with on the same pad but different color paletettes?
This is possible, but requires a touch of fancy ROOT-ing. Actually, it's not that much, it just takes a lot of time to figure it out. So here it is.
Fair warning, the order of all the gPad->Modified's etc seems very important to no seg fault.
In include the main (demo.cc) and the Makefile. |
| Attachment 1: demo.cc
|
///////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
/////// demo.cxx
////// Nov 2018, Brian Clark
////// Demonstrate how to draw 2 TH2D's with two different color palettes (needs ROOT6)
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
//C++
#include <iostream>
//ROOT Includes
#include "TH2D.h"
#include "TCanvas.h"
#include "TStyle.h"
#include "TExec.h"
#include "TColor.h"
#include "TPaletteAxis.h"
#include "TRandom3.h"
using namespace std;
int main(int argc, char **argv)
{
gStyle->SetOptStat(0);
TH2D *h1 = new TH2D("h1","h1",100,-10,10,100,-10,10);
TH2D *h2 = new TH2D("h2","h2",100,-10,10,100,-10,10);
TRandom3 *R = new TRandom3(time(0));
int count=0;
while(count<10000000){
h1->Fill(R->Gaus(-4.,1.),R->Gaus(-4.,1.));
count++;
}
TRandom3 *R2 = new TRandom3(time(0)+100);
count=0;
while(count<10000000){
h2->Fill(R2->Gaus(4.,1.),R2->Gaus(4.,1.));
count++;
}
TCanvas *c = new TCanvas("c","c",1100,850);
h1->Draw("colz");
TExec *ex1 = new TExec("ex1","gStyle->SetPalette(kValentine);");
ex1->Draw();
h1->Draw("colz same");
gPad->SetLogz();
gPad->Modified();
gPad->Update();
TPaletteAxis *palette1 = (TPaletteAxis*)h1->GetListOfFunctions()->FindObject("palette");
palette1->SetX1NDC(0.7);
palette1->SetX2NDC(0.75);
palette1->SetY1NDC(0.1);
palette1->SetY2NDC(0.95);
gPad->SetRightMargin(0.3);
gPad->SetTopMargin(0.05);
h2->Draw("same colz");
TExec *ex2 = new TExec("ex2","gStyle->SetPalette(kCopper);"); //TColor::InvertPalette();
ex2->Draw();
h2->Draw("same colz");
gPad->SetLogz();
gPad->Modified();
gPad->Update();
TPaletteAxis *palette2 = (TPaletteAxis*)h2->GetListOfFunctions()->FindObject("palette");
palette2->SetX1NDC(0.85);
palette2->SetX2NDC(0.90);
palette2->SetY1NDC(0.1);
palette2->SetY2NDC(0.95);
h1->SetTitle("");
h1->GetXaxis()->SetTitle("X-Value");
h1->GetYaxis()->SetTitle("Y-Value");
h1->GetZaxis()->SetTitle("First Histogram Events");
h2->GetZaxis()->SetTitle("Second Histogram Events");
h1->GetYaxis()->SetTitleSize(0.05);
h1->GetXaxis()->SetTitleSize(0.05);
h1->GetYaxis()->SetLabelSize(0.045);
h1->GetXaxis()->SetLabelSize(0.045);
h1->GetZaxis()->SetTitleSize(0.045);
h2->GetZaxis()->SetTitleSize(0.045);
h1->GetZaxis()->SetLabelSize(0.04);
h2->GetZaxis()->SetLabelSize(0.04);
h1->GetYaxis()->SetTitleOffset(1);
h1->GetZaxis()->SetTitleOffset(1);
h2->GetZaxis()->SetTitleOffset(1);
c->SetLogz();
c->SaveAs("test.png");
}
|
| Attachment 2: Makefile
|
LDFLAGS=-L${ARA_UTIL_INSTALL_DIR}/lib -L${shell root-config --libdir}
CXXFLAGS=-I${ARA_UTIL_INSTALL_DIR}/include -I${shell root-config --incdir}
LDLIBS += $(shell $(ROOTSYS)/bin/root-config --libs)
|
| Attachment 3: test.png
|  |
|
|
32
|
Mon Dec 17 21:16:31 2018 |
Brian Clark | Run over many data files in parallel | | To analyze data, we sometimes need to run over many thousands of runs at once. To do this in parallel, we can submit a job for every run we want to do. This will proceed in several steps:
- We need to prepare an analysis program.
- This is demo.cxx.
- The program will take an input data file and an output location.
- The program will do some analysis on each events, and then write the result of that analysis to an output file labeled by the same number as the input file.
- We need to prepare a job script for PBS.
- This is "run.sh"; this is the set of instructions to be submitted to the cluster.
- The instructions say to:
- Source a a shell environment
- To run the executable
- Move the output root file to the output location.
- Note that we're telling the program we wrote in step 1 to write to the node-local $TMPDIR, and then moving the result to our final output directory at the end. This is better for cluster performance.
- We need to make a list of data files to run over
- We can do this on OSC by running
ls -d -1 /fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event*.root > run_list.txt
- This places the full path to the ROOT files in that folder into a list called run_list.txt that we can loop over.
- Third, we need to script that will submit all of the jobs to the cluster.
- This is "submit_jobs.sh".
- This loops over all the files in our run_list.txt and submits a run.sh job for each of them.
- This is also where we define the $RUNDIR (where the code is to be exeucted) and the $OUTPUTDIR (where the output products are to be stored)
Once you've generated all of these output files, you can run over the output files only to make plots and such.
|
| Attachment 1: demo.cxx
|
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
//// demo.cxx
//// demo
////
//// Nov 2018
////////////////////////////////////////////////////////////////////////////////
//Includes
#include <iostream>
#include <string>
#include <sstream>
//AraRoot Includes
#include "RawAtriStationEvent.h"
#include "UsefulAtriStationEvent.h"
//ROOT Includes
#include "TTree.h"
#include "TFile.h"
#include "TGraph.h"
using namespace std;
RawAtriStationEvent *rawAtriEvPtr;
int main(int argc, char **argv)
{
if(argc<3) {
std::cout << "Usage\n" << argv[0] << " <input_file> <output_location> "<<endl;
return -1;
}
/*
arguments
0: exec
1: input data file
2: output location
*/
TFile *fpIn = TFile::Open(argv[1]);
if(!fpIn) {
std::cout << "Can't open file\n";
return -1;
}
TTree *eventTree = (TTree*) fpIn->Get("eventTree");
if(!eventTree) {
std::cout << "Can't find eventTree\n";
return -1;
}
eventTree->SetBranchAddress("event",&rawAtriEvPtr);
int run;
eventTree->SetBranchAddress("run",&run);
eventTree->GetEntry(0);
printf("Filter Run Number %d \n", run);
char outfile_name[400];
sprintf(outfile_name,"%s/outputs_run%d.root",argv[2],run);
TFile *fpOut = TFile::Open(outfile_name, "RECREATE");
TTree* outTree = new TTree("outTree", "outTree");
int WaveformLength[16];
outTree->Branch("WaveformLength", &WaveformLength, "WaveformLength[16]/D");
Long64_t numEntries=eventTree->GetEntries();
for(Long64_t event=0;event<numEntries;event++) {
eventTree->GetEntry(event);
UsefulAtriStationEvent *realAtriEvPtr = new UsefulAtriStationEvent(rawAtriEvPtr, AraCalType::kLatestCalib);
for(int i=0; i<16; i++){
TGraph *gr = realAtriEvPtr->getGraphFromRFChan(i);
WaveformLength[i] = gr->GetN();
delete gr;
}
outTree->Fill();
delete realAtriEvPtr;
} //loop over events
fpOut->Write();
fpOut->Close();
fpIn->Close();
delete fpIn;
}
|
| Attachment 2: run.sh
|
#/bin/bash
#PBS -l nodes=1:ppn=1
#PBS -l mem=4GB
#PBS -l walltime=00:05:00
#PBS -A PAS0654
#PBS -e /fs/scratch/PAS0654/shell_demo/err_out_logs
#PBS -o /fs/scratch/PAS0654/shell_demo/err_out_logs
# you should change the -e and -o to write your
# log files to a location of your preference
# source your own shell script here
eval 'source /users/PAS0654/osu0673/A23_analysis/env.sh'
# $RUNDIR was defined in the submission script
# along with $FILE and $OUTPUTDIR
cd $RUNDIR
# $TMPDIR is the local memory of this specific node
# it's the only variable we didn't have to define
./bin/demo $FILE $TMPDIR
# after we're done
# we copy the results to the $OUTPUTDIR
pbsdcp $TMPDIR/'*.root' $OUTPUTDIR
|
| Attachment 3: run_list.txt
|
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1000.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1001.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1002.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1004.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1005.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1006.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1007.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1009.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1010.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1011.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1012.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1014.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1015.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1016.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1017.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1019.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1020.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1021.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1022.root
/fs/scratch/PAS0654/ara/10pct/RawData/A3/2013/sym_links/event1024.root
|
| Attachment 4: submit_jobs.sh
|
#!/bin/bash
#where should the outputs be stored?
OutputDir="/fs/scratch/PAS0654/shell_demo/outputs"
echo '[ Processed file output directory: ' $OutputDir ' ]'
export OutputDir
#where is your executable compiled?
RunDir="/users/PAS0654/osu0673/A23_analysis/araROOT"
export RunDir
#define the list of runs to execute on
readfile=run_list.txt
counter=0
while read line1
do
qsub -v RUNDIR=$RunDir,OUTPUTDIR=$OutputDir,FILE=$line1 -N 'job_'$counter run.sh
counter=$((counter+1))
done < $readfile
|
|
|
33
|
Mon Feb 11 21:58:26 2019 |
Brian Clark | Get a quick start with icemc on OSC | Software | Follow the instructions in the attached "getting_started_with_anita.pdf" file to download icemc, compile it, generate results, and plot those results. |
| Attachment 1: sample_bash_profile.sh
|
# .bash_profile
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
# User specific environment and startup programs
PATH=$PATH:$HOME/.local/bin:$HOME/bin
export PATH
|
| Attachment 2: sample_bashrc.sh
|
# .bashrc
source bashrc_anita.sh
# we also want to set two more environment variables that ANITA needs
# you should update ICEMC_SRC_DIR and ICEMC_BUILD_DIR to wherever you
# downloaded icemc too
export ICEMC_SRC_DIR=/path/to/icemc #change this line!
export ICEMC_BUILD_DIR=/path/to/icemc #change this line!
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
|
| Attachment 3: test_plot.cc
|
//C++ includes
#include <iostream>
//ROOT includes
#include "TCanvas.h"
#include "TStyle.h"
#include "TH1D.h"
#include "TFile.h"
#include "TTree.h"
using namespace std;
int main(int argc, char *argv[])
{
if(argc<2)
{
cout << "Not enough arguments! Stop run. " << endl;
return -1;
}
/*
we're going to make a histogram, and set some parameters about it's X and Y axes
*/
TH1D *nuflavorint_hist = new TH1D("nuflavorint", "",3,1,4);
nuflavorint_hist->SetTitle("Neutrino Flavors");
nuflavorint_hist->GetXaxis()->SetTitle("Neutrino Flavors (1=e, 2=muon, 3=tau)");
nuflavorint_hist->GetYaxis()->SetTitle("Weigthed Fraction of Total Detected Events");
nuflavorint_hist->GetXaxis()->SetTitleOffset(1.2);
nuflavorint_hist->GetYaxis()->SetTitleOffset(1.2);
nuflavorint_hist->GetXaxis()->CenterTitle();
nuflavorint_hist->GetYaxis()->CenterTitle();
for(int i=1; i < argc; i++)
{ // loop over the input files
//now we are going to load the icefinal.root file and draw in the "passing_events" tree, which stores info
string readfile = string(argv[i]);
TFile *AnitaFile = new TFile(( readfile ).c_str());
cout << "AnitaFile" << endl;
TTree *passing_events = (TTree*)AnitaFile->Get("passing_events");
cout << "Reading AnitaFile..." << endl;
//declare three variables we are going to use later
int num_pass; // number of entries (ultra-neutrinos);
double weight; // weight of neutrino counts;
int nuflavorint; // neutrino flavors;
num_pass = passing_events->GetEntries();
cout << "num_pass is " << num_pass << endl;
/*PRIMARIES VARIABLES*/
//set the "branch" of the tree which stores specific pieces of information
passing_events->SetBranchAddress("weight", &weight);
passing_events->SetBranchAddress("nuflavor", &nuflavorint);
//loop over all the events in the tree
for (int k=0; k <=num_pass; k++)
{
passing_events->GetEvent(k);
nuflavorint_hist->Fill(nuflavorint, weight); //fill the histogram with this value and this weight
} // CLOSE FOR LOOP OVER NUMBER OF EVENTS
} // CLOSE FOR LOOP OVER NUMBER OF INPUT FILES
//set up some parameters to make things loo pretty
gStyle->SetHistFillColor(0);
gStyle->SetHistFillStyle(1);
gStyle->SetHistLineColor(1);
gStyle->SetHistLineStyle(0);
gStyle->SetHistLineWidth(2.5); //Setup plot Style
//make a "canvas" to draw on
TCanvas *c4 = new TCanvas("c4", "nuflavorint", 1100,850);
gStyle->SetOptTitle(1);
gStyle->SetStatX(0.33);
gStyle->SetStatY(0.87);
nuflavorint_hist->Draw("HIST"); //draw on it
//Save Plots
//make the line thicker and then save the result
gStyle->SetHistLineWidth(9);
c4->SaveAs("nuflavorint.png");
gStyle->SetHistLineWidth(2);
c4->SaveAs("nuflavorint.pdf");
delete c4; //clean up
return 0; //return successfully
}
|
| Attachment 4: bashrc_anita.sh
|
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
module load cmake/3.11.4
module load gnu/7.3.0
export CC=`which gcc`
export CXX=`which g++`
export BOOST_ROOT=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/boost_build
export LD_LIBRARY_PATH=${BOOST_ROOT}/stage/lib:$LD_LIBRARY_PATH
export BOOST_LIB=$BOOST_ROOT/stage/lib
export LD_LIBRARY_PATH=$BOOST_LIB:$LD_LIBRARY_PATH
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
eval 'source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh'
|
| Attachment 5: getting_running_with_anita_stuff.pdf
|
| Attachment 6: getting_running_with_anita_stuff.pptx
|
| Attachment 7: test_plot.mk
|
# Makefile for the ROOT test programs. # This Makefile shows nicely how to compile and link applications
# using the ROOT libraries on all supported platforms.
#
# Copyright (c) 2000 Rene Brun and Fons Rademakers
#
# Author: Fons Rademakers, 29/2/2000
include Makefile.arch
################################################################################
# Site specific flags
################################################################################
# Toggle these as needed to get things to install
#BOOSTFLAGS = -I boost_1_48_0
# commented out for kingbee and older versions of gcc
ANITA3_EVENTREADER=1
# Uncomment to enable healpix
#USE_HEALPIX=1
# Uncomment to disable explicit vectorization (but will do nothing if ANITA_UTIL is not available)
#VECTORIZE=1
# The ROOT flags are added to the CXXFLAGS in the .arch file
# so this should be simpler...
ifeq (,$(findstring -std=, $(CXXFLAGS)))
ifeq ($(shell test $(GCC_MAJOR) -lt 5; echo $$?),0)
ifeq ($(shell test $(GCC_MINOR) -lt 5; echo $$?),0)
CXXFLAGS += -std=c++0x
else
CXXFLAGS += -std=c++11
endif
endif
endif
################################################################################
# If not compiling with C++11 (or later) support, all occurrences of "constexpr"
# must be replaced with "const", because "constexpr" is a keyword
# which pre-C++11 compilers do not support.
# ("constexpr" is needed in the code to perform in-class initialization
# of static non-integral member objects, i.e.:
# static const double c_light = 2.99e8;
# which works in C++03 compilers, must be modified to:
# static constexpr double c_light = 2.99e8;
# to work in C++11, but adding "constexpr" breaks C++03 compatibility.
# The following compiler flag defines a preprocessor macro which is
# simply:
# #define constexpr const
# which replaces all instances of the text "constexpr" and replaces it
# with "const".
# This preserves functionality while only affecting very specific semantics.
ifeq (,$(findstring -std=c++1, $(CXXFLAGS)))
CPPSTD_FLAGS = -Dconstexpr=const
endif
# Uses the standard ANITA environment variable to figure
# out if ANITA libs are installed
ifdef ANITA_UTIL_INSTALL_DIR
ANITA_UTIL_EXISTS=1
ANITA_UTIL_LIB_DIR=${ANITA_UTIL_INSTALL_DIR}/lib
ANITA_UTIL_INC_DIR=${ANITA_UTIL_INSTALL_DIR}/include
LD_ANITA_UTIL=-L$(ANITA_UTIL_LIB_DIR)
LIBS_ANITA_UTIL=-lAnitaEvent -lRootFftwWrapper
INC_ANITA_UTIL=-I$(ANITA_UTIL_INC_DIR)
ANITA_UTIL_ETC_DIR=$(ANITA_UTIL_INSTALL_DIR)/etc
endif
ifdef ANITA_UTIL_EXISTS
CXXFLAGS += -DANITA_UTIL_EXISTS
endif
ifdef VECTORIZE
CXXFLAGS += -DVECTORIZE -march=native -fabi-version=0
endif
ifdef ANITA3_EVENTREADER
CXXFLAGS += -DANITA3_EVENTREADER
endif
ifdef USE_HEALPIX
CXXFLAGS += -DUSE_HEALPIX `pkg-config --cflags healpix_cxx`
LIBS += `pkg-config --libs healpix_cxx`
endif
################################################################################
GENERAL_FLAGS = -g -O2 -pipe -m64 -pthread
WARN_FLAGS = -W -Wall -Wextra -Woverloaded-virtual
# -Wno-unused-variable -Wno-unused-parameter -Wno-unused-but-set-variable
CXXFLAGS += $(GENERAL_FLAGS) $(CPPSTD_FLAGS) $(WARN_FLAGS) $(ROOTCFLAGS) $(INC_ANITA_UTIL)
DBGFLAGS = -pipe -Wall -W -Woverloaded-virtual -g -ggdb -O0 -fno-inline
DBGCXXFLAGS = $(DBGFLAGS) $(ROOTCFLAGS) $(BOOSTFLAGS)
LDFLAGS += $(CPPSTD_FLAGS) $(LD_ANITA_UTIL) -I$(BOOST_ROOT) -L.
LIBS += $(LIBS_ANITA_UTIL)
# Mathmore not included in the standard ROOT libs
LIBS += -lMathMore
DICT = classdict
OBJS = vector.o position.o earthmodel.o balloon.o icemodel.o signal.o ray.o Spectra.o anita.o roughness.o secondaries.o Primaries.o Tools.o counting.o $(DICT).o Settings.o Taumodel.o screen.o GlobalTrigger.o ChanTrigger.o SimulatedSignal.o EnvironmentVariable.o source.o random.o
BINARIES = test_plot$(ExeSuf)
################################################################################
.SUFFIXES: .$(SrcSuf) .$(ObjSuf) .$(DllSuf)
all: $(BINARIES)
$(BINARIES): %: %.$(SrcSuf) $(OBJS)
$(LD) $(CXXFLAGS) $(LDFLAGS) $(OBJS) $< $(LIBS) $(OutPutOpt) $@
@echo "$@ done"
.PHONY: clean
clean:
@rm -f $(BINARIES)
%.$(ObjSuf) : %.$(SrcSuf) %.h
@echo "<**Compiling**> "$<
$(LD) $(CXXFLAGS) -c $< -o $@
|
|
|
38
|
Wed May 15 00:38:54 2019 |
Brian Clark | Get a quick start with AraSim on osc | Software | Follow the instructions in the attached "getting_started_with_ara.pdf" file to download AraSim, compile it, generate results, and plot those results. |
| Attachment 1: plotting_example.cc
|
#include <iostream>
#include <fstream>
#include <sstream>
#include <math.h>
#include <string>
#include <stdio.h>
#include <stdlib.h>
#include <vector>
#include <time.h>
#include <cstdio>
#include <cstdlib>
#include <unistd.h>
#include <cstring>
#include <unistd.h>
#include "TTreeIndex.h"
#include "TChain.h"
#include "TH1.h"
#include "TF1.h"
#include "TF2.h"
#include "TFile.h"
#include "TRandom.h"
#include "TRandom2.h"
#include "TRandom3.h"
#include "TTree.h"
#include "TLegend.h"
#include "TLine.h"
#include "TROOT.h"
#include "TPostScript.h"
#include "TCanvas.h"
#include "TH2F.h"
#include "TText.h"
#include "TProfile.h"
#include "TGraphErrors.h"
#include "TStyle.h"
#include "TMath.h"
#include "TVector3.h"
#include "TRotation.h"
#include "TSpline.h"
#include "Math/InterpolationTypes.h"
#include "Math/Interpolator.h"
#include "Math/Integrator.h"
#include "TGaxis.h"
#include "TPaveStats.h"
#include "TLatex.h"
#include "Constants.h"
#include "Settings.h"
#include "Position.h"
#include "EarthModel.h"
#include "Tools.h"
#include "Vector.h"
#include "IceModel.h"
#include "Trigger.h"
#include "Spectra.h"
#include "signal.hh"
#include "secondaries.hh"
#include "Ray.h"
#include "counting.hh"
#include "Primaries.h"
#include "Efficiencies.h"
#include "Event.h"
#include "Detector.h"
#include "Report.h"
using namespace std;
/////////////////Plotting Script for nuflavorint for AraSIMQC
/////////////////Created by Kaeli Hughes, modified by Brian Clark
/////////////////Prepared on April 28 2016 as an "introductor script" to plotting with AraSim; for new users of queenbee
class EarthModel;
int main(int argc, char **argv)
{
gStyle->SetOptStat(111111); //this is a selection of statistics settings; you should do some googling and figure out exactly what this particular comibation does
gStyle->SetOptDate(1); //this tells root to put a date and timestamp on whatever plot we output
if(argc<2){ //if you don't have at least 1 file to run over, then you haven't given it a file to analyze; this checks this
cout<<"Not enough arguments! Abort run."<<endl;
}
//Create the histogram
TCanvas *c2 = new TCanvas("c2", "nuflavorint", 1100,850); //make a canvas on which to plot the data
TH1F *nuflavorint_hist = new TH1F("nuflavorint_hist", "nuflavorint histogram", 3, 0.5, 3.5); //create a histogram with three bins
nuflavorint_hist->GetXaxis()->SetNdivisions(3); //set the number of divisions
int total_thrown=0; // a variable to hold the grant total number of events thrown for all input files
for(int i=1; i<argc;i++){ //loop over the input files
string readfile; //create a variable called readfile that will hold the title of the simulation file
readfile = string(argv[i]); //set the readfile variable equal to the filename
Event *event = 0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
Report *report=0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
Settings *settings = 0; //create a Event class pointer called event; note that it is set equal to zero to avoid creating a bald pointer
TFile *AraFile=new TFile(( readfile ).c_str()); //make a new file called "AraFile" that will be simulation file we are reading in
if(!(AraFile->IsOpen())) return 0; //checking to see if the file we're trying to read in opened correctly; if not, bounce out of the program
int num_pass;//number of passing events
TTree *AraTree=(TTree*)AraFile->Get("AraTree"); //get the AraTree
TTree *AraTree2=(TTree*)AraFile->Get("AraTree2"); //get the AraTree2
AraTree2->SetBranchAddress("event",&event); //get the event branch
AraTree2->SetBranchAddress("report",&report); //get the report branch
AraTree->SetBranchAddress("settings",&settings); //get the settings branch
num_pass=AraTree2->GetEntries(); //get the number of passed events in the data file
AraTree->GetEvent(0); //get the first event; sometimes the tree does not instantiate properly if you'd explicitly "active" the first event
total_thrown+=(settings->NNU); //add the number of events from this file (The NNU variable) to the grand total; NNU is the number of THROWN neutrinos
for (int k=0; k<num_pass; k++){ //going to fill the histograms for as many events as were in this input file
AraTree2->GetEvent(k); //get the even from the tree
int nuflavorint; //make the container variable
double weight; //the weight of the event
int trigger; //the global trigger value for the event
nuflavorint=event->nuflavorint; //draw the event out; one of the objects in the event class is the nuflavorint, and this is the syntax for accessing it
weight=event->Nu_Interaction[0].weight; //draw out the weight of the event
trigger=report->stations[0].Global_Pass; //find out if the event was a triggered event or not
/*
if(trigger!=0){ //check if the event triggered
nuflavorint_hist->Fill(nuflavorint,weight); //fill the event into the histogram; the first argument of the fill (which is mandatory) is the value you're putting into the histogram; the second value is optional, and is the weight of the event in the histogram
//in this particular version of the code then, we are only plotting the TRIGGERED events; if you wanted to plot all of the events, you could instead remove this "if" condition and just Fill everything
}
*/
nuflavorint_hist->Fill(nuflavorint,weight); //fill the event into the histogram; the first argument of the fill (which is mandatory)
}
}
//After looping over all the files, make the plots and save them
//do some stuff to get the "total events thrown" text box ready; this is useful because on the plot itself you can then see how many events you THREW into the simulation; this is especially useful if you're only plotting passed events, but want to know what fraction of your total thrown that is
char *buffer= (char*) malloc (250); //declare a buffer pointer, and allocate it some chunk of memory
int a = snprintf(buffer, 250,"Total Events Thrown: %d",total_thrown); //print the words "Total Events Thrown: %d" to the variable "buffer" and tell the system how long that phrase is; the %d sign tells C++ to replace that "%d" with the next argument, or in this case, the number "total_thrown"
if(a>=250){ //if the phrase is longer than the pre-allocated space, increase the size of the buffer until it's big enough
buffer=(char*) realloc(buffer, a+1);
snprintf(buffer, 250,"Total Events Thrown: %d",total_thrown);
}
TLatex *u = new TLatex(.3,.01,buffer); //create a latex tex object which we can draw on the cavas
u->SetNDC(kTRUE); //changes the coordinate system for the tex object plotting
u->SetIndiceSize(.1); //set the size of the latex index
u->SetTextSize(.025); //set the size of the latex text
nuflavorint_hist->Draw(); //draw the histogram
nuflavorint_hist->GetXaxis()->SetTitle("Neutrino Flavor"); //set the x-axis label
nuflavorint_hist->GetYaxis()->SetTitle("Number of Events (weighted)"); //set the y-axis label
nuflavorint_hist->GetYaxis()->SetTitleOffset(1.5); //set the separation between the y-axis and it's label; root natively makes this smaller than is ideal
nuflavorint_hist->SetLineColor(kBlack); //set the color of the histogram to black, instead of root's default navy blue
u->Draw(); //draw the statistics box information
c2->SaveAs("outputs/plotting_example.png"); //save the canvas as a JPG file for viewing
c2->SaveAs("outputs/plotting_example.pdf"); //save the canvas as a PDF file for viewing
c2->SaveAs("outputs/plotting_example.root"); //save the canvas as a ROOT file for viewing or editing later
} //end main; this is the end of the script
|
| Attachment 2: plotting_example.mk
|
#############################################################################
## Makefile -- New Version of my Makefile that works on both linux
## and mac os x
## Ryan Nichol <rjn@hep.ucl.ac.uk>
##############################################################################
##############################################################################
##############################################################################
##
##This file was copied from M.readGeom and altered for my use 14 May 2014
##Khalida Hendricks.
##
##Modified by Brian Clark for use on plotting_example on 28 April 2016
##
##Changes:
##line 54 - OBJS = .... add filename.o .... del oldfilename.o
##line 55 - CCFILE = .... add filename.cc .... del oldfilename.cc
##line 58 - PROGRAMS = filename
##line 62 - filename : $(OBJS)
##
##
##############################################################################
##############################################################################
##############################################################################
include StandardDefinitions.mk
#Site Specific Flags
ifeq ($(strip $(BOOST_ROOT)),)
BOOST_ROOT = /usr/local/include
endif
SYSINCLUDES = -I/usr/include -I$(BOOST_ROOT)
SYSLIBS = -L/usr/lib
DLLSUF = ${DllSuf}
OBJSUF = ${ObjSuf}
SRCSUF = ${SrcSuf}
CXX = g++
#Generic and Site Specific Flags
CXXFLAGS += $(INC_ARA_UTIL) $(SYSINCLUDES)
LDFLAGS += -g $(LD_ARA_UTIL) -I$(BOOST_ROOT) $(ROOTLDFLAGS) -L.
# copy from ray_solver_makefile (removed -lAra part)
# added for Fortran to C++
LIBS = $(ROOTLIBS) -lMinuit $(SYSLIBS)
GLIBS = $(ROOTGLIBS) $(SYSLIBS)
LIB_DIR = ./lib
INC_DIR = ./include
#ROOT_LIBRARY = libAra.${DLLSUF}
OBJS = Vector.o EarthModel.o IceModel.o Trigger.o Ray.o Tools.o Efficiencies.o Event.o Detector.o Position.o Spectra.o RayTrace.o RayTrace_IceModels.o signal.o secondaries.o Settings.o Primaries.o counting.o RaySolver.o Report.o eventSimDict.o plotting_example.o
CCFILE = Vector.cc EarthModel.cc IceModel.cc Trigger.cc Ray.cc Tools.cc Efficiencies.cc Event.cc Detector.cc Spectra.cc Position.cc RayTrace.cc signal.cc secondaries.cc RayTrace_IceModels.cc Settings.cc Primaries.cc counting.cc RaySolver.cc Report.cc plotting_example.cc
CLASS_HEADERS = Trigger.h Detector.h Settings.h Spectra.h IceModel.h Primaries.h Report.h Event.h secondaries.hh #need to add headers which added to Tree Branch
PROGRAMS = plotting_example
all : $(PROGRAMS)
plotting_example : $(OBJS)
$(LD) $(OBJS) $(LDFLAGS) $(LIBS) -o $(PROGRAMS)
@echo "done."
#The library
$(ROOT_LIBRARY) : $(LIB_OBJS)
@echo "Linking $@ ..."
ifeq ($(PLATFORM),macosx)
# We need to make both the .dylib and the .so
$(LD) $(SOFLAGS)$@ $(LDFLAGS) $(G77LDFLAGS) $^ $(OutPutOpt) $@
ifneq ($(subst $(MACOSX_MINOR),,1234),1234)
ifeq ($(MACOSX_MINOR),4)
ln -sf $@ $(subst .$(DllSuf),.so,$@)
else
$(LD) -dynamiclib -undefined $(UNDEFOPT) $(LDFLAGS) $(G77LDFLAGS) $^ \
$(OutPutOpt) $(subst .$(DllSuf),.so,$@)
endif
endif
else
$(LD) $(SOFLAGS) $(LDFLAGS) $(G77LDFLAGS) $(LIBS) $(LIB_OBJS) -o $@
endif
##-bundle
#%.$(OBJSUF) : %.$(SRCSUF)
# @echo "<**Compiling**> "$<
# $(CXX) $(CXXFLAGS) -c $< -o $@
%.$(OBJSUF) : %.C
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
%.$(OBJSUF) : %.cc
@echo "<**Compiling**> "$<
$(CXX) $(CXXFLAGS) $ -c $< -o $@
# added for fortran code compiling
%.$(OBJSUF) : %.f
@echo "<**Compiling**> "$<
$(G77) -c $<
eventSimDict.C: $(CLASS_HEADERS)
@echo "Generating dictionary ..."
@ rm -f *Dict*
rootcint $@ -c ${INC_ARA_UTIL} $(CLASS_HEADERS) ${ARA_ROOT_HEADERS} LinkDef.h
clean:
@rm -f *Dict*
@rm -f *.${OBJSUF}
@rm -f $(LIBRARY)
@rm -f $(ROOT_LIBRARY)
@rm -f $(subst .$(DLLSUF),.so,$(ROOT_LIBRARY))
@rm -f $(TEST)
#############################################################################
|
| Attachment 3: test_setup.txt
|
NFOUR=1024
EXPONENT=21
NNU=300 // number of neutrino events
NNU_PASSED=10 // number of neutrino events that are allowed to pass the trigger
ONLY_PASSED_EVENTS=0 // 0 (default): AraSim throws NNU events whether or not they pass; 1: AraSim throws events until the number of events that pass the trigger is equal to NNU_PASSED (WARNING: may cause long run times if reasonable values are not chosen)
NOISE_WAVEFORM_GENERATE_MODE=0 // generate new noise waveforms for each events
NOISE_EVENTS=16 // number of pure noise waveforms
TRIG_ANALYSIS_MODE=0 // 0 = signal + noise, 1 = signal only, 2 = noise only
DETECTOR=1 // ARA stations 1 to 7
NOFZ=1
core_x=10000
core_y=10000
TIMESTEP=5.E-10 // value for 2GHz actual station value
TRIG_WINDOW=1.E-7 // 100ns which is actual testbed trig window
POWERTHRESHOLD=-6.06 // 100Hz global trig rate for 3 out of 16 ARA stations
POSNU_RADIUS=3000
V_MIMIC_MODE=0 // 0 : global trig is located center of readout windows
DATA_SAVE_MODE=0 // 2 : don't save any waveform informations at all
DATA_LIKE_OUTPUT=0 // 0 : don't save any waveform information to eventTree
BORE_HOLE_ANTENNA_LAYOUT=0
SECONDARIES=0
TRIG_ONLY_BH_ON=0
CALPULSER_ON=0
USE_MANUAL_GAINOFFSET=0
USE_TESTBED_RFCM_ON=0
NOISE_TEMP_MODE=0
TRIG_THRES_MODE=0
READGEOM=0 // reads geometry information from the sqlite file or not (0 : don't read)
TRIG_MODE=0 // use vpol, hpol separated trigger mode. by default N_TRIG_V=3, N_TRIG_H=3. You can change this values
number_of_stations=1
core_x=10000
core_y=10000
DETECTOR=1
DETECTOR_STATION=2
DATA_LIKE_OUTPUT=0
NOISE_WAVEFORM_GENERATE_MODE=0 // generates new waveforms for every event
NOISE=0 //flat thermal noise
NOISE_CHANNEL_MODE=0 //using different noise temperature for each channel
NOISE_EVENTS=16 // number of noise events which will be store in the trigger class for later use
ANTENNA_MODE=1
APPLY_NOISE_FIGURE=0
|
| Attachment 4: bashrc_anita.sh
|
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
module load cmake/3.11.4
module load gnu/7.3.0
export CC=`which gcc`
export CXX=`which g++`
export BOOST_ROOT=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/boost_build
export LD_LIBRARY_PATH=${BOOST_ROOT}/stage/lib:$LD_LIBRARY_PATH
export BOOST_LIB=$BOOST_ROOT/stage/lib
export LD_LIBRARY_PATH=$BOOST_LIB:$LD_LIBRARY_PATH
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
eval 'source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh'
|
| Attachment 5: sample_bash_profile.sh
|
# .bash_profile
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
# User specific environment and startup programs
PATH=$PATH:$HOME/.local/bin:$HOME/bin
export PATH
|
| Attachment 6: sample_bashrc.sh
|
# .bashrc
source bashrc_anita.sh
# we also want to set two more environment variables that ANITA needs
# you should update ICEMC_SRC_DIR and ICEMC_BUILD_DIR to wherever you
# downloaded icemc too
export ICEMC_SRC_DIR=/path/to/icemc #change this line!
export ICEMC_BUILD_DIR=/path/to/icemc #change this line!
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
|
| Attachment 7: getting_running_with_ara_stuff.pdf
|
| Attachment 8: getting_running_with_ara_stuff.pptx
|
|
|
25
|
Wed Aug 1 11:37:10 2018 |
Andres Medina | Flux Ordering | Hardware | Bought 951 Non-Resin Soldering Flux. This is the preferred variety. This could be found on this website https://www.kester.com/products/product/951-soldering-flux.
The amount of Flux that was bought was 1 Gallon (Lasts quite some time). The price was $83.86 with approximate shipping of $43. This was done with a Pcard and a tax exempt form.
The website used to purchase this was https://www.alliedelec.com/kester-solder-63-0000-0951/70177935/ |
|
|
12
|
Fri Aug 25 12:34:47 2017 |
Amy Connolly - posting stuff from Todd Thompson | How to run coffee | Other | One really useful thing in here is how to describe "Value Added" to visitors. |
| Attachment 1: guidelines.pdf
|
|
|
1
|
Thu Mar 16 09:01:50 2017 |
Amy Connolly | Elog instructions | Other | Log into kingbee.mps.ohio-state.edu first, then log into radiorm.physics.ohio-state.edu.
From Keith Stewart 03/16/17: It appears that radiorm SSH from offsite is closed. So you will need to be on an OSU network physically or via VPN. fox is also blocked from offsite as well. Kingbee should still be available for now. If you want to use it as a jump host to get to radiorm without VPN. However, you will want to get comfortable with the VPN before it is a requirement.
Carl 03/16/17: I could log in even while using a hard line and plugged in directly to the network.
From Bryan Dunlap 12/16/17: I have set up the group permissions on the elog directory so you and your other designated people can edit files. I have configured sudo to allow you all to restart the elogd service. Once you have edited the file [/home/elog/elog.cfg I think], you can then type
sudo /sbin/service elogd restart
to restart the daemon so it re-reads the config. Sudo will prompt you for your password before it executes the command. |
|
|
2
|
Thu Mar 16 10:39:15 2017 |
Amy Connolly | How Do I Connect to the ASC VPN Using Cisco and Duo? | | For Mac and Windows:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=14542
For Linux, in case some of your students need it:
https://osuasc.teamdynamix.com/TDClient/KB/ArticleDet?ID=17908
From Sam 01/25/17: It doesn't work from my Ubuntu 14 machine. My VPN setup in 14 does not have the "Software Token Authentication" option on the screen as shown in the instructions. It fails on connection attempt.
The instructions specify Ubuntu 16; perhaps there is a way to make it work on 14, but I don't know what it is.
|
|
|
5
|
Tue Apr 18 12:02:55 2017 |
Amy Connolly | How to ship to Pole | Hardware | Here is an old email thread about how to ship a station to Pole.
|
| Attachment 1: Shipping_stuff_to_Pole__a_short_how_to_from_you_would_be_nice_.pdf
|
| Attachment 2: ARA_12-13_UH_to_CHC_Packing_List_Box_2_of_3.pdf
|
| Attachment 3: ARA_12-13_UH_to_CHC_Packing_List_Box_1_of_3.pdf
|
| Attachment 4: ARA_12-13_UH_to_CHC_Packing_List_Box_3_of_3.pdf
|
| Attachment 5: IMG_4441.jpg
|  |
|
|
37
|
Tue May 14 10:38:08 2019 |
Amy | Getting started with AraSim | Software | Attached is a set of slides on Getting Started with QC, a simulation monitoring project. It has instructions on getting started in using a terminal window, and downloading, compiling and running AraSim, the simulation program for the ARA project. AraSim has moved from the SVN repository to github, and so now you should be able to retrieve it, compile it using:
git clone https://github.com/ara-software/AraSim.git
cd AraSim
make
./AraSim
It will run without arguments, but the output might be silly. You can follow the instructions for running AraSim that are in the qc_Intro instructions, which will give them not silly results. Those parts are still correct.
You might get some const expr errors if you are using ROOT 6, such as the ones in the first screen grab below. As mentioned in the error messages, you need to change from const expr to not. A few examples are shown in the next screen grab.
If you are here, you likely would also want to know how to install the prerequisites themselves. You might find this entry helpful then: http://radiorm.physics.ohio-state.edu/elog/How-To/4. It is only technically applicable to an older version that is designed for compatibility with ROOT5, but it will give you the idea.
These instructions are also superceded by an updated presentation at http://radiorm.physics.ohio-state.edu/elog/How-To/38
|
| Attachment 1: intro_to_qc.pdf
|
| Attachment 2: Screenshot_from_2019-05-14_10-54-48.png
|  |
| Attachment 3: Screenshot_from_2019-05-14_10-56-19.png
|  |
|
|
41
|
Fri Oct 11 13:43:35 2019 |
Amy | How to start a new undergrad hire | Other | If an undergrad has been working with the group on a volunteer basis, they will need that clarified, so that they have not up to then we working for pay without training. There is a form that they sign saying they have been working as a volunteer.
Below is an email that Pam sent in July 2019 outlining what they need. Other things to remember:
Undergrads receive emails from the department to remind them about orientation and scheduling prior to first day of hire, and other emails from ASC. They receive more information at orientation.
They will need to show ID.
If an undergrad (or any hire) does not waive retirement/OPERs within 30 days, they will have to have this deducted from their paycheck. This is another reason that orientation is important.
For the person hiring them:
1. Tell your admin (Lisa) asap who, when, etc you want to hire. ASAP is important because (Pam) will process over 75 undergraduates hires in the Fall and over 100 in the summer. Each takes 20 days on average (see below)
2. On the first day of employment – ask your new hire if they have completed their orientation with the ASC.
From: Hood, Pam
Sent: Tuesday, July 23, 2019 4:49 PM
To: 'physics-all@lists.osu.edu' <physics-all@lists.osu.edu>
Subject: Undergraduate Fall Hires
Hello all,
Fall is almost upon us and I am working on having positions ready to fill as well as posting on the OSU student job site and Federal Work study job boards. In order to plan my workflow, if you would let me know:
1. Approximately how many undergraduate student assistants you plan to hire and at what rate of pay. Our department’s current average rate of pay is $10.00 - $10.50/hour, however it does depend on the position. Range is $8.55 - $14.00/hour.
2. A brief summary of job duties and responsibilities (i.e. assisting in research for ______)
3. Request for posting OR
4. If you have specific student that has been a non-paid volunteer that you would like to hire, they need to sign a waiver prior to orientation. Please see attached for volunteer waiver.
5. Please indicate if your start date varies from August 20. And it may vary based on multiply factors i.e. signatures, workflow, etc.
6. All terminations for summer ungraduated student workers OR
7. If you are intending to continue employment but require a reduction in hours worked in order to comply with policy Please see attached policy for min/max hours.
I have attempted to answer all the FAQs that I noted in the “summer undergraduate hiring wave” in italics, however please let me know if you have questions. As always, it is ASC policy to not start any employee prior to orientation and it will take a minimum of 10 days from the time the contract is signed by both the student and the supervisor– please encourage your students to sign asap or it lengthens the process significantly.
I will do my best to facilitate this process for you. Students Buck ID# and email is extremely helpful. If you need the hire to start August 20, please send me the information via email by July 31, 2019. If you respond later or request a hire after July 31, I will estimate the hire date at the time of submittal.
Thanks,
Pam |
|
|
46
|
Tue Aug 2 14:34:15 2022 |
Alex M | OSC License Request | | Some programs on OSC require authorized access to use in the form of a license. The license will be automatically read if it is available whenever you open a program on OSC, provided you have access to the license. In order to have access to a license, you need to fill out the attached form and send it to Amy to forward to OSC. Available programs (at least some of) which require a license can be found here: https://www.osc.edu/resources/available_software/software_list . Replace the name of the program at the top of the form with the desired software. |
| Attachment 1: User_Software_Agreement.pdf
|
|
|
48
|
Thu Jun 8 16:29:45 2023 |
Alan Salcedo | Doing IceCube/ARA coincidence analysis | | These documents contain information on how to run IceCube/ARA coincidence simulations and analysis. All technical information of where codes are stored and how to use them is detailed in the technical note. Other supportive information for physics understanding is in the powerpoint slides. The technical note will direct you to other documents in this elog in the places where you may need supplemental information. |
| Attachment 1: IceCube_ARA_Coincidence_Analysis___Technical_Note.pdf
|
| Attachment 2: ICARA_Coincident_Events_Introduction.pptx
|
| Attachment 3: ICARA_Analysis_Template.ipynb
|
{
"cells": [
{
"cell_type": "markdown",
"id": "bcdfb138",
"metadata": {},
"source": [
"# IC/ARA Coincident Simulation Events Analysis"
]
},
{
"cell_type": "markdown",
"id": "c7ad7c80",
"metadata": {},
"source": [
"### Settings and imports"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "b6915a86",
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"<style>.container { width:75% !important; }</style>"
],
"text/plain": [
"<IPython.core.display.HTML object>"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"## Makes this notebook maximally wide\n",
"from IPython.display import display, HTML\n",
"display(HTML(\"<style>.container { width:75% !important; }</style>\"))"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "6ef9e20b",
"metadata": {},
"outputs": [],
"source": [
"## Author: Alex Machtay (machtay.1@osu.edu)\n",
"## Modified by: Alan Salcedo (salcedogomez.1@osu.edu)\n",
"## Date: 4/26/23\n",
"\n",
"## Purpose:\n",
"### This script will read the data files produced by AraRun_corrected_MultStat.py to make histograms relevant plots of\n",
"### neutrino events passing through icecube and detected by ARA station (in AraSim)\n",
"\n",
"\n",
"## Imports\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"import sys\n",
"sys.path.append(\"/users/PAS0654/osu8354/root6_18_build/lib\") # go to parent dir\n",
"sys.path.append(\"/users/PCON0003/cond0068/.local/lib/python3.6/site-packages\")\n",
"import math\n",
"import argparse\n",
"import glob\n",
"import pandas as pd\n",
"pd.options.mode.chained_assignment = None # default='warn'\n",
"from mpl_toolkits.mplot3d import Axes3D\n",
"import jupyterthemes as jt"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "f24b8292",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"/bin/bash: jt: command not found\r\n"
]
}
],
"source": [
"## Set style for the jupyter notebook\n",
"!jt -t grade3 -T -N -kl -lineh 160 -f code -fs 14 -ofs 14 -cursc o\n",
"jt.jtplot.style('grade3', gridlines='')"
]
},
{
"cell_type": "markdown",
"id": "a5a812d6",
"metadata": {},
"source": [
"### Set constants\n",
"#### These are things like the position of ARA station holes, the South Pole, IceCube's position in ARA station's coordinates, and IceCube's radius"
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "cdb851de",
"metadata": {},
"outputs": [],
"source": [
"## What IceCube (IC) station are you analyzing\n",
"station = 1\n",
"\n",
"## What's the radius around ARA where neutrinos were injected\n",
"inj_rad = 5 #in km\n",
"\n",
"## IceCube's center relative to each ARA station\n",
"IceCube = [[-1128.08, -2089.42, -1942.39], [-335.812, -3929.26, -1938.23],\n",
" [-2320.67, -3695.78, -1937.35], [-3153.04, -1856.05, -1942.81], [472.49, -5732.98, -1922.06]] #IceCube's position relative to A1, A2, or A3\n",
"\n",
"#To calculate this, we need to do some coordinate transformations. Refer to this notebook to see the calculations: \n",
"# IceCube_Relative_to_ARA_Stations.ipynb (found here - http://radiorm.physics.ohio-state.edu/elog/How-To/48) \n",
"\n",
"## IceCube's radius\n",
"IceCube_radius = 564.189583548 #Modelling IceCube as a cylinder, we find the radius with V = h * pi*r^2 with V = 1x10^9 m^3 and h = 1x10^3 m "
]
},
{
"cell_type": "markdown",
"id": "af5c6fc2",
"metadata": {},
"source": [
"### Read the data\n",
"\n",
"#### Once we import the data, we'll make dataframes to concatenate it and make some calculations"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "921f61f8",
"metadata": {},
"outputs": [],
"source": [
"## Import data files\n",
"\n",
"#Here, it's from OSC Connolly's group project space\n",
"source = '/fs/project/PAS0654/IceCube_ARA_Coincident_Search/AraSim/outputs/Coincident_Search_Runs/20M_GZK_5km_S1_correct' \n",
"num_files = 200 # Number of files to read in from the source directory\n",
"\n",
"## Make a list of all of the paths to check \n",
"file_list = []\n",
"for i in range(1, num_files + 1):\n",
" for name in glob.glob(source + \"/\" + str(i) + \"/*.csv\"):\n",
" file_list.append(str(name))\n",
" #file_list gets paths to .csv files\n",
" \n",
"## Now read the csv files into a pandas dataframe\n",
"dfs = []\n",
"for filename in file_list:\n",
" df = pd.read_csv(filename, index_col=None, header=0) #Store each csv file into a pandas data frame\n",
" dfs.append(df) #Append the csv file to store all of them in one\n",
"frame = pd.concat(dfs, axis=0, ignore_index = True) #Concatenate pandas dataframes "
]
},
{
"cell_type": "markdown",
"id": "813fb3ee",
"metadata": {},
"source": [
"### Work with the data\n",
"\n",
"#### All the data from our coincidence simulations (made by AraRun_MultStat.sh) is now stored in a pandas data frame that we can work with"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "8b449583",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"19800000\n",
"19799802\n"
]
}
],
"source": [
"## Now let's clean up our data and calculate other relevant things\n",
"print(len(frame))\n",
"frame = frame[frame['weight'].between(0,1)] #Filter out events with ill-defined weight (should be between 0 and 1)\n",
"print(len(frame))\n",
"\n",
"frame['x-positions'] = (frame['Radius (m)'] * np.cos(frame['Azimuth (rad)']) * np.sin(frame['Zenith (rad)']))\n",
"frame['y-positions'] = (frame['Radius (m)'] * np.sin(frame['Azimuth (rad)']) * np.sin(frame['Zenith (rad)']))\n",
"frame['z-positions'] = (frame['Radius (m)'] * np.cos(frame['Zenith (rad)']))\n",
"\n",
"## The energy in eV will be 10 raised to the number in the file, multiplied by 1-y (y is inelasticity)\n",
"frame['Nu Energies (eV)'] = np.power(10, (frame['Energy (log10) (eV)']))\n",
"frame['Mu Energies (eV)'] = ((1-frame['Inelasticity']) * frame['Nu Energies (eV)']) #Energy of the produced lepton\n",
"#Here the lepton is not a muon necesarily, hence the label 'Mu Energies (eV)' may be misleading\n",
"\n",
"## Get a frame with only coincident events\n",
"coincident_frame = frame[frame['Coincident'] == 1] \n",
"\n",
"## And a frame for strictly events *detected* by ARA\n",
"detected_frame = frame[frame['Detected'] == 1]\n",
"\n",
"\n",
"## Now let's calculate the energy of the lepton when reaching IceCube (IC)\n",
"\n",
"# To do this correctly, I need to find exactly the distance traveled by the muon and apply the equation\n",
"# I need the trajectory of the muon to find the time it takes to reach IceCube, then I can find the distance it travels in that time\n",
"# I should allow events that occur inside the icecube volume to have their full energy (but pretty much will happen anyway)\n",
"## a = sin(Theta)*cos(Phi)\n",
"## b = sin(Theta)*sin(Phi)\n",
"## c = cos(Theta)\n",
"## a_0 = x-position\n",
"## b_0 = y-position\n",
"## c_0 = z-position\n",
"## x_0 = IceCube[0]\n",
"## y_0 = IceCube[1]\n",
"## z_0 = IceCube[2]\n",
"## t = (-(a*(a_0-x_0) + b*(b_0-y_0))+D**0.5)/(a**2+b**2)\n",
"## D = (a**2+b**2)*R_IC**2 - (a*(b_0-y_0)+b*(a_0-x_0))**2\n",
"## d = ((a*t)**2 + (b*t)**2 + (c*t)**2)**0.5\n",
"\n",
"## Trajectories\n",
"coincident_frame['a'] = (np.sin(coincident_frame['Theta (rad)'])*np.cos(coincident_frame['Phi (rad)']))\n",
"coincident_frame['b'] = (np.sin(coincident_frame['Theta (rad)'])*np.sin(coincident_frame['Phi (rad)']))\n",
"coincident_frame['c'] = (np.cos(coincident_frame['Theta (rad)']))\n",
"\n",
"## Discriminant\n",
"coincident_frame['D'] = ((coincident_frame['a']**2 + coincident_frame['b']**2)*IceCube_radius**2 - \n",
" (coincident_frame['a']*(coincident_frame['y-position (m)']-IceCube[station-1][1])- ## I think this might need to be a minus sign!\n",
" coincident_frame['b']*(coincident_frame['x-position (m)']-IceCube[station-1][0]))**2)\n",
"\n",
"## Interaction time (this is actually the same as the distance traveled, at least for a straight line)\n",
"coincident_frame['t_1'] = (-(coincident_frame['a']*(coincident_frame['x-position (m)']-IceCube[station-1][0])+\n",
" coincident_frame['b']*(coincident_frame['y-position (m)']-IceCube[station-1][1]))+\n",
" np.sqrt(coincident_frame['D']))/(coincident_frame['a']**2+coincident_frame['b']**2)\n",
"coincident_frame['t_2'] = (-(coincident_frame['a']*(coincident_frame['x-position (m)']-IceCube[station-1][0])+\n",
" coincident_frame['b']*(coincident_frame['y-position (m)']-IceCube[station-1][1]))-\n",
" np.sqrt(coincident_frame['D']))/(coincident_frame['a']**2+coincident_frame['b']**2)\n",
"\n",
"## Intersection coordinates\n",
"coincident_frame['x-intersect_1'] = (coincident_frame['a'] * coincident_frame['t_1'] + coincident_frame['x-position (m)'])\n",
"coincident_frame['y-intersect_1'] = (coincident_frame['b'] * coincident_frame['t_1'] + coincident_frame['y-position (m)'])\n",
"coincident_frame['z-intersect_1'] = (coincident_frame['c'] * coincident_frame['t_1'] + coincident_frame['z-position (m)'])\n",
"\n",
"coincident_frame['x-intersect_2'] = (coincident_frame['a'] * coincident_frame['t_2'] + coincident_frame['x-position (m)'])\n",
"coincident_frame['y-intersect_2'] = (coincident_frame['b'] * coincident_frame['t_2'] + coincident_frame['y-position (m)'])\n",
"coincident_frame['z-intersect_2'] = (coincident_frame['c'] * coincident_frame['t_2'] + coincident_frame['z-position (m)'])\n",
"\n",
"## Distance traveled (same as the parametric time, at least for a straight line)\n",
"coincident_frame['d_1'] = (np.sqrt((coincident_frame['a']*coincident_frame['t_1'])**2+\n",
" (coincident_frame['b']*coincident_frame['t_1'])**2+\n",
" (coincident_frame['c']*coincident_frame['t_1'])**2))\n",
"coincident_frame['d_2'] = (np.sqrt((coincident_frame['a']*coincident_frame['t_2'])**2+\n",
" (coincident_frame['b']*coincident_frame['t_2'])**2+\n",
" (coincident_frame['c']*coincident_frame['t_2'])**2))\n",
"\n",
"## Check if it started inside and set the distance based on if it needs to travel to reach icecube or not\n",
"coincident_frame['Inside'] = (np.where((coincident_frame['t_1']/coincident_frame['t_2'] < 0) & (coincident_frame['z-position (m)'].between(-2450, -1450)), 1, 0))\n",
"coincident_frame['preliminary d'] = (np.where(coincident_frame['d_1'] <= coincident_frame['d_2'], coincident_frame['d_1'], coincident_frame['d_2']))\n",
"coincident_frame['d'] = (np.where(coincident_frame['Inside'] == 1, 0, coincident_frame['preliminary d']))\n",
"\n",
"## Check if the event lies in the cylinder\n",
"coincident_frame['In IC'] = (np.where((np.sqrt((coincident_frame['x-position (m)']-IceCube[station-1][0])**2 + (coincident_frame['y-position (m)']-IceCube[station-1][1])**2) < IceCube_radius) &\n",
" ((coincident_frame['z-position (m)']).between(-2450, -1450)) , 1, 0))\n",
"\n",
"#Correct coincident_frame to only have electron neutrinos inside IC\n",
"coincident_frame = coincident_frame[(((coincident_frame['In IC'] == 1) & (coincident_frame['flavor'] == 1)) | (coincident_frame['flavor'] == 2) | (coincident_frame['flavor'] == 3)) ]\n",
"\n",
"#Now calculate the lepton energies when they reach IC\n",
"coincident_frame['IC Mu Energies (eV)'] = (coincident_frame['Mu Energies (eV)'] * np.exp(-10**-5 * coincident_frame['d']*100)) # convert d from meters to cm\n",
"coincident_frame['weighted energies'] = (coincident_frame['weight'] * coincident_frame['Nu Energies (eV)'])"
]
},
{
"cell_type": "code",
"execution_count": 7,
"id": "f1757103",
"metadata": {
"scrolled": false
},
"outputs": [],
"source": [
"## Add possible Tau decay to the frame\n",
"coincident_frame['Tau decay'] = ''\n",
"# Again, the label 'Tau Decay' may be misleading because not all leptons may be taus\n",
"\n",
"## Calculate distance from the interaction point to its walls and keep the shortest (the first interaction with the volume)\n",
"\n",
"coincident_frame['distance-to-IC_1'] = np.sqrt((coincident_frame['x-positions'] - coincident_frame['x-intersect_1'])**2 + \n",
" (coincident_frame['y-positions'] - coincident_frame['y-intersect_1'])**2)\n",
"coincident_frame['distance-to-IC_2'] = np.sqrt((coincident_frame['x-positions'] - coincident_frame['x-intersect_2'])**2 + \n",
... 19001 more lines ...
|
| Attachment 4: IceCube_Relative_to_ARA_Stations.ipynb
|
{
"cells": [
{
"cell_type": "markdown",
"id": "a950b9af",
"metadata": {},
"source": [
"**This script is simply for me to calculate the location of IceCube relative to the origin of any ARA station**\n",
"\n",
"The relevant documentation to understand the definitions after the imports can be found in https://elog.phys.hawaii.edu/elog/ARA/130712_170712/doc.pdf"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "b926e2e3",
"metadata": {},
"outputs": [],
"source": [
"import numpy as np"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "901b442b",
"metadata": {},
"outputs": [],
"source": [
"#Definitions of translations in surveyor's coordinates:\n",
"\n",
"t_IC_to_ARAg = np.array([-24100, 1700, 6400])\n",
"t_ARAg_to_A1 = np.array([16401.71, -2835.37, -25.67])\n",
"t_ARAg_to_A2 = np.array([13126.7, -8519.62, -18.72])\n",
"t_ARAg_to_A3 = np.array([9848.35, -2835.19, -12.7])\n",
"\n",
"#Definitions of rotations from surveyor's axes to the ARA Station's coordinate systems\n",
"\n",
"R1 = np.array([[-0.598647, 0.801013, -0.000332979], [-0.801013, -0.598647, -0.000401329], \\\n",
" [-0.000520806, 0.0000264661, 1]])\n",
"R2 = np.array([[-0.598647, 0.801013, -0.000970507], [-0.801007, -0.598646,-0.00316072 ], \\\n",
" [-0.00311277, -0.00111477, 0.999995]])\n",
"R3 = np.array([[-0.598646, 0.801011, -0.00198193],[-0.801008, -0.598649,-0.00247504], \\\n",
" [-0.00316902, 0.000105871, 0.999995]])"
]
},
{
"cell_type": "markdown",
"id": "ab2d3206",
"metadata": {},
"source": [
"**Using these definitions, I should be able to calculate the location of IceCube relative to each ARA station by:**\n",
"\n",
"$$\n",
"\\vec{r}_{A 1}^{I C}=-R_1\\left(\\vec{t}_{I C}^{A R A}+\\vec{t}_{A R A}^{A 1}\\right)\n",
"$$\n",
"\n",
"We have a write-up of how to get this. Contact salcedogomez.1@osu.edu if you need that.\n",
"\n",
"Alex had done this already, he got that \n",
"\n",
"$$\n",
"\\vec{r}_{A 1}^{I C}=-3696.99^{\\prime} \\hat{x}-6843.56^{\\prime} \\hat{y}-6378.31^{\\prime} \\hat{z}\n",
"$$\n",
"\n",
"Let me verify that I get the same"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "912163d2",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A1 (in): [-3696.98956579 -6843.55800868 -6378.30926681]\n",
"IC coordinates relative to A1 (m): [-1127.13096518 -2086.4506124 -1944.60648378]\n",
"Distance of IC from A1 (m): 3066.788996234438\n"
]
}
],
"source": [
"IC_A1 = -R1 @ np.add(t_ARAg_to_A1, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A1 (in): \", IC_A1)\n",
"print(\"IC coordinates relative to A1 (m): \", IC_A1/3.28)\n",
"print(\"Distance of IC from A1 (m): \", np.sqrt((IC_A1[0]/3.28)**2 + (IC_A1[1]/3.28)**2 + (IC_A1[2]/3.28)**2))"
]
},
{
"cell_type": "markdown",
"id": "f9c9f252",
"metadata": {},
"source": [
"Looks good!\n",
"\n",
"Now, I just get the other ones:"
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "8afa27c6",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A2 (in): [ -1100.33577313 -12852.0589083 -6423.00776043]\n",
"IC coordinates relative to A2 (m): [ -335.46822352 -3918.31064277 -1958.2340733 ]\n",
"Distance of IC from A2 (m): 4393.219537890439\n"
]
}
],
"source": [
"IC_A2 = -R2 @ np.add(t_ARAg_to_A2, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A2 (in): \", IC_A2)\n",
"print(\"IC coordinates relative to A2 (m): \", IC_A2/3.28)\n",
"print(\"Distance of IC from A2 (m): \", np.sqrt((IC_A2[0]/3.28)**2 + (IC_A2[1]/3.28)**2 + (IC_A2[2]/3.28)**2))"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "9959d0a4",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"IC coordinates relative to A3 (in): [ -7609.73440732 -12079.45719852 -6432.31164368]\n",
"IC coordinates relative to A3 (m): [-2320.04097784 -3682.76134101 -1961.07062307]\n",
"Distance of IC from A3 (m): 4774.00452685144\n"
]
}
],
"source": [
"IC_A3 = -R3 @ np.add(t_ARAg_to_A3, t_IC_to_ARAg).T\n",
"print(\"IC coordinates relative to A3 (in): \", IC_A3)\n",
"print(\"IC coordinates relative to A3 (m): \", IC_A3/3.28)\n",
"print(\"Distance of IC from A3 (m): \", np.sqrt((IC_A3[0]/3.28)**2 + (IC_A3[1]/3.28)**2 + (IC_A3[2]/3.28)**2))"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "093dff67",
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.12"
}
},
"nbformat": 4,
"nbformat_minor": 5
}
|
|
|
4
|
Fri Mar 31 11:36:54 2017 |
Brian Clark, Hannah Hasan, Jude Rajasekera, and Carl Pfendner | Installing Software Pre-Requisites for Simulation and Analysis | Software | Instructions on installing simulation software prerequisites (ROOT, Boost, etc) on Linux computers. |
| Attachment 1: installation.pdf
|
| Attachment 2: Installation-Instructions.tar.gz
|
|