| |
ID |
Date |
Author |
Subject |
|
|
183
|
Mon Oct 24 16:07:22 2022 |
Dylan Wells | Creating the matching circuit design | The first part of designing the matching crictuit was choosing a frequency range to match over. We chose 100 MHz to 1000 MHz.
The geometric mean of this range in 316.227 MHz, which is the ideal frequeny to match to.
Then, we need the impedance of the antenna we are matching to.
The data for the best antenna is found in
/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/AraSim_Polarity_Fix_2021_03_19/AraSim_Polarity_Fix_2021_03_19.xf/Simulations/001108/Run0001/output/SteadyStateOutput
The files begin at 83.33 MHz and increment by 16.66 MHz, so file 14 (316.57 MHz) is the best choice for matching.
This antenna has Impedance Z = 229.839 -i151.515 ohms
From here, we need to decide a design architecture for the matching circuit. Since the real component of the impedance is greater than the source's (50 ohms, the standard for electrical sources), a natural design is a lowpass downward circuit.
Next, we decide on a number of L networks to cascade in the circuit design, or how many 'rungs' in the ladder.
The purpose of these rungs is to artificially lower the ratio between the source impedance and the load impedance, as the closer these values, the broader the range of frequencies we can match.
The formula to calculate this is
N = ln(Rl/Rs)/ln(1+Q^2)
where
Q = ((Rl/Rs)-1)^½
Rs = real source impedance
Rl = real load impedance
For the given values, N is 14.
From here, we have all the information to find the capacitances and impedances for each L network in the ladder circuit.
The proccess of calciulating these values starts with finding the desired ratio between the impedances.
ratio = Q^2+1
Which we can use to create a list of 13 impedances between the source and load (corresponding to 14 rungs), each Q^2+1 times larger than the last. Then we can use the formula for a single lowpass downward match between the impedances of each rung.
All of this process is described in /users/PAS1960/dylanwells1629/improved_match_maker.py
The key functions PCSL, calcN, and broadbandMatchLP
|
|
|
182
|
Fri Sep 2 12:40:17 2022 |
Alan S | Ice Shell Around Bicone | Here is a PDF with a summary/log of my work trying to integrate an ice shell surrounding the bicone antennas in the GENETIS loop. I made gain patterns of the ARA bicone with the ice shells and without them, showing the impact of the ice on the antenna's gain. Having an ice shell changes these patterns, which are also dependent on the radius of the shell. I showed that convergence on the gain patterns and expected features start ocurring for shells with radii larger than 10 meters. I also keep track of the computation time which increases with bigger shells.
Note: I also add PowerPoint slides that contain all the graphs that I made. The PDF does not contain some of those since I didn't think they were coherent with my write-up, but I may be making an omission.
|
| Attachment 1: XF_ARA_bicone_in_ice_and_air_column.pptx
|
| Attachment 2: Far_Zone_Gain_of_ARA_Bicone_insice_Ice.pdf
|
|
|
181
|
Thu Aug 18 13:03:47 2022 |
Dylan Wells | Comparing inputs and outputs between AraSim and IceMC | Comparing Inputs:
Frequency Lists:
Ara - 83.33MHz - 1066.70 MHz, step = 13.33MHz
IceMC - 200MHz - 1500MHz, step = 10MHz
Number of files read in:
Ara - 1
IceMC - 8
Formating of files:
Ara - Theta, Phi, Gain (dB, thetra), Gain (theta), Phase (theta)
IceMc - Frequency, Gain (dB) (Different files cover different thetas and phis)
File Type:
Ara - .txt
IceMC - no suffix (file with 2 columns of text)
Other notes for IceMC inputs:
IceMC reads in 8 different files for gain.
vv_0 hh_0 vh_0 hv_0 vv_el vv_az hh_el hh_az
Found in ../anitaBuiltTool/components/icemc/data
vv_0 = gains for vertical polarization
hh_0 = gains for horizontal polarization
vh_0 = gains for v → h cross polarization
hv_0 = gains for h → v cross polarization
vv_el = v polarization, e angle
vv_az = v polarization, a angle
hh_el = h polarization, e angle
hh_az = h polarization a angle
for e angle and a angle in
0. 0
-
5
-
10
-
20
-
30
-
45
-
90
(iterates 1 to 6)
Comparing Outputs:
IceMC - veff is in second column in veff+runName+.txt file (in the outputDirectory directory)
Ara - veff is at the bottom of the AraOut.txt file |
|
|
180
|
Thu Aug 18 10:32:17 2022 |
Amy | reading about HPol antennas | Here are some reading materials about HPol antennas:
This paper discusses some initial HPol designs that were tested in a prototype station of ARA. We went with the slotted cylinder design.
Here you can see what the RNO-G experiment in Greenland has considered.
We can add other sources here as we find them. |
|
|
179
|
Tue Aug 16 11:41:07 2022 |
Dylan Wells | Instructions for Running IceMC | Running IceMC:
Go into the directory ../anitaBuildTool/build/components/icemc/
And run the command
*May need to chmod -R 775 ../anitaBuildTool/comonents/icemc/ if you get a permissions error
inputFile:
Must be the full path to the file
Config files are found in ../anitaBuildTool/components/icemc
Ex: /users/PAS1960/dylanwells1629/anitaBuildTool/components/icemc/inputs.anita4.conf
Config files are found in ../anitaBuildTool/components/icemc
outputDirectory:
Will be made in ../anitaBuildTool/build/components/icemc/ by default, specify full path otherwise.
runNumber:
The run number.
numberOfNeutrinos:
The number of neutrinos generated in the simulation.
Can be found in inputs.conf
Default is 2,000,000.
#How many neutrinos to generate
triggerThreshold:
Threshold for each band for the trigger.
Default is 2.3
#thresholds for each band- this is only for the frequency domain voltage trigger. If using a different trigger scheme then keep these at the default values of 2.3 because the max among them is used for the chance in hell cuts
energyExponent:
The exponent of the energy for the neutrinos
Can be found in input.conf
Default is 1020
# Select energy (just enter the exponent) or (30) for baseline ES&S (1) for E^-1 (2) for E^-2 (3) for E^-3 (4) for E^-4 (5) for ES&S flux with cosmological constant (6) for neutrino GZK flux from Iron nuclei (16-22)not using spectrum but just for a single energy (101-114)use all kinds of theoretical flux models |
|
|
178
|
Wed Aug 10 22:38:20 2022 |
Dylan Wells | Instructions for Installing IceMC | Installing IceMC:
I: Getting anitaBuildTool
Clone the anitaBuildTool repository (https://github.com/anitaNeutrino/anitaBuildTool) to your user.
II: Get the Anita.sh file onto your user.
Either copy the file from /users/PAS1960/dylanwells1629/Anita.sh
or create a new file Anita.sh with the following code
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
#this modules were originally loading in both the env.sh, and bashrc_anita.sh files. This was redundant so it was added here, and removed from the others.
module load gnu/7.3.0
module load gnu
module load mvapich2
module load fftw3
#module load python/3.6-conda5.2
module load cmake
PATH=$PATH:$HOME/.local/bin:$home/bin
export PATH
export CC=`which gcc`
export CXX=`which g++`
export FFTWDIR=/fs/project/PAS0654/shared_software/fftw3/gnu/6.3/mvapich2/2.2/3.3.5
export ANITA_SOURCE_DIR=~/anitaBuildTool/
export ANITA_UTIL_INSTALL_DIR=~/anitaBuildTool/
export ICEMC_SRC_DIR=~/anitaBuildTool/components/icemc/
export ICEMC_BUILD_DIR=~/anitaBuildTool/build/components/icemc/
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
# User specific aliases and functions
#This env.sh is for running the BiconeEvolution GENETIS software. This should only be un-commented if you are running GENETIS software. When you do this, comment out env.sh.
#source ~/new_root_setup.sh
source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh
#source /cvmfs/ara.opensciencegrid.org/trunk/centos7/setup.sh
#module load python/3.6-conda5.2
#BiconeGENETIS directory shortcut SHARED
alias GE='cd ../../../fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/'
#emacs Alias
alias emacs='emacs -nw'
#root alias
alias root='root -l'
#Alias
alias l="ls"
alias python='/cvmfs/ara.opensciencegrid.org/trunk/centos7/misc_build/bin/python3.9'
Then source the file
Note: You will need access to PAS0654 for this step or you will get a permissions error.
III: Running the build tool.
Go into the anitaBuildTool directory
And run the building script
Note: There will be an error if you source files for running Ara in your .bashrc
Comment these out and restart your terminal before running the build. (remember to source Anita.sh before running the build tool. You could also source Anita.sh in your .bashrc)

Error if you source files for running Ara:
CMake Error at components/libRootFftwWrapper/cmake_install.cmake:238 (file):
file INSTALL cannot copy file
"/users/PAS1960/dylanwells1629/anitaBuildTool/components/libRootFftwWrapper/include/AnalyticSignal.h"
to
"/cvmfs/ara.opensciencegrid.org/v2.0.0/centos7/ara_build/include/AnalyticSignal.h":
Read-only file system.
Call Stack (most recent call first):
|
|
|
177
|
Tue Aug 9 15:28:50 2022 |
Ryan Debolt | How many individuals to use in the GA. | One of our foundational questions tied to the optimization of the GA has been "How many individuals should we simulate". Up to now, our minds were made up for us by the speed of arasim being great enough that the time cost of simulating individuals was great enough that the improvements made from having more were not enough to justify the slowdown. However, with the upgrade to the faster, more recent version of arasim, I decided to re-examine this. This was also spurred on by the fact that the last time I ran this test we were testing GA performance by final generation metrics rather than by how many generations it took to reach a benchmark. So in one of my optimization tests, I tracked this data.
To start, using the same run proportions, using a .5 chi-squared benchmark, the average time across all 89 run types used in this run was 25.4 generations for 50 individuals as compared to 8.3 generations running the same test for 100. Furthermore, the minimum number of generations for 50 individuals was 4.8 while using 100 individuals yielded 2.4. So on average running 100 individuals was about 3 times fast at reaching this benchmark than with 50. And when comparing the best result regardless of run type, 100 individuals was still 2 times quicker than the min for 50 individuals. Finally, the run that yielded an average of 2.4 generations for 100 individuals took an average of 29.2 generations with 50 individuals or roughly 12 times the generations.
For the test we will discuss, we ran 89 different run types that all used 60% rank, 20% roulette, and 20% tournament selection respectively. These test had the following ranges:
6-18% of individuals through reproduction (steps of 3%)
64-88% of individuals through crossover (steps of 12%)
0-10% mutation rate (steps of 5%)
1-5% sigma on mutation (steps of 1%)
These tests also used our fitness scores with simulated error of .1 to imitate arasim's behavior and as such we used the chi-squared value to evaluate these scores as there is no error on those values.
Comparing this same test with a tighter chi-squared benchmark of .25, we see similar results. On average 50 individuals took 37.1 generations to reach this point while 100 individuals took 16.0 generations. Similarly, the minimums amount of gens for 50 individuals was 15.4 while 100 individuals was 5. Finally, the corresponding run for the 5 generation min with 100 individuals took 41.8 generations with 50 individuals. These correspond to speed up's of 2.3, 3.08, and 8.36 respectively.
This data implies that on average, independent of run type, we should expect to have to use 2-3 times fewer generations while running 100 individuals than we would running 50 individuals but we could see up to 8-12 times fewer generations to reach benchmarks. Another data set using a different set of selection methods was also tested for this and again yielded similar results, though overall the runs from the first batch were better across both 50 and 100 individuals and so those results are likely to be more indicative of the parameters we use in a true run.
The data being examined in these results can be found here: https://docs.google.com/spreadsheets/d/1GlfnjQSO6VI8MuUGYTUcLkjwDZU98nyFFysgTTfVFOE/edit?usp=sharing |
|
|
176
|
Tue Aug 9 11:36:24 2022 |
Ryan Debolt | GA User guide (pdf) | |
| Attachment 1: VPol_GA_User_guide.pdf
|
|
|
175
|
Fri Aug 5 09:59:07 2022 |
Dennis Calderon | Updated Effective Volumes and Errors (Paper Run and Curved Sides) | Update to previous ELOG post for Effective Volumes and Errors for AraSim simulations for antennas from Paper_Run and Curved_Sides.
AraSim version compiled ~11/2021 and the (old) GENETIS version
Here are the column definitions:
- The first column indicates the antenna being described. For example, XF_model_21_20 was the 20th individual in the 21st generation for the run discussed in the paper. Curved_side_1 was the best performing individual from the curved run done in February 2022. Curved_Side_1_Quad_Zero indicates the curved individual with its quadratic term set to 0. Curved_Side_1_Straightened indicates the curved individual with the quadratic term set to zero and the linear term modified so as to keep the outer radius the same as it originally was (that is, for Curved_Side_1).
- The next three columns are the measurements for that individual.
- The first one idicates the mean effective volume of the runs done for that individual. There were ~300 runs, each of 10,000 neutrinos.
- The second column indicates the standard deviation of the mean (the previous column). This was measured by taking the standard deviation of the effective volumes (the line that says test Veff(ice) in the AraOut file) from all 300 runs (that is, Sigma(mean - x_i)/Sqrt(300)).
- example of line:
- Veff(ice) : 5.25337e+09 m3sr, 5.25337 km3sr
- The final column indicates the average of the errors on the effective volume measurements from the AraOut files (the line that Veff(water eq.)) divided by Sqrt(300) for each of the 300 jobs (that is, Sigma(x_i/Sqrt(300))/300).
- example of line:
- And Veff(water eq.) error plus : 0.552722 and error minus : 0.552722
Note: Previously, the best curved individual (according to the run data) had been excluded (instead, we had the 2nd-6th best individuals). This has been corrected, with the order corrected.
Also find attached a file called full_resimulate_DNA.csv . It contains the DNA for the top five curved individuals, the DNA for those same individuals with their quadratic term set to 0, and the DNA for those individuals with the quadratic term set to zero and the linear term modified to maintain the same outer radius as in the original run.
Finally, the three images show the original best curved bicone (Curved_Side_1), the best curved bicone when its quadratic gene was set to 0 (Curved_Side_1_Quad_Zero), and the best curved bicone when its quadratic gene was set to 0 *and* its linear gene was changed to keep the same outer radius (Curved_Side_1_Straightened).
We have chosen XF_Model_23_08 to be the one actually built for testing. This was decided based on the data presented here. XF_Model_23_08 was the highest scoring individual in the paper run. Because the paper data was generated using the old version of AraSim, it was decided that using those results would make the most sense for which antenna to build. This would also mean that we can compare directly to the best individual in the paper. This individual was chosen over the curved sided inviduals because it has a higher fitness score, both when using the old version of AraSim and the new version. |
| Attachment 1: Effective_Volumes_Errors_2.pdf
|
| Attachment 2: full_resimulate_DNA.csv
|
Hybrid of Roulette and Tournament -- Thanks to Cal Poly / Jordan Potter
Author was David Liu
Notable contributors: Julie Rolla, Hannah Hasan, and Adam Blenk
Done at The Ohio State University
Working on behalf of Dr. Amy Connolly
And the ANITA project
Revision date: 21 March 2018 1800 EST
0.08333,0.1,0.11667,0.13334,0.15001,0.16668,0.18335,0.20002,0.21669,0.23336,0.25003,0.2667,0.28337,0.30004,0.31671,0.33338,0.35005,0.36672,0.38339,0.40006,0.41673,0.4334,0.45007,0.46674,0.48341,0.50008,0.51675,0.53342,0.55009,0.56676,0.58343,0.6001,0.61677,0.63344,0.65011,0.66678,0.68345,0.70012,0.71679,0.73346,0.75013,0.7668,0.78347,0.80014,0.81681,0.83348,0.85015,0.86682,0.88349,0.90016,0.91683,0.9335,0.95017,0.96684,0.98351,1.00018,1.01685,1.03352,1.05019,1.06686
Matrices for this Generation:
6.31483,77.6017,0.00260615,-0.197494
3.97024,40.9244,-0.0103313,0.36119
2.32499,79.663,-0.00224948,0.192602
0.308809,39.6742,-0.0084247,0.40629
2.32499,75.9855,-0.00224948,0.192602
0.966617,42.4246,-0.0084247,0.40629
5.83731,79.663,-0.000594616,0.0106887
2.316,38.0641,0.00166678,0.0135261
4.66148,79.663,-0.000594616,0.00582814
1.75822,39.9608,-0.000356019,0.0167485
6.31483,77.6017,0.0,-0.197494
3.97024,40.9244,0.0,0.36119
2.32499,79.663,0.0,0.192602
0.308809,39.6742,0.0,0.40629
2.32499,75.9855,0.0,0.192602
0.966617,42.4246,0.0,0.40629
5.83731,79.663,0.0,0.0106887
2.316,38.0641,0.0,0.0135261
4.66148,79.663,0.0,0.00582814
1.75822,39.9608,0.0,0.0167485
6.31483,77.6017,0.0,0.00474767
3.97024,40.9244,0.0,-0.0616123
2.32499,79.663,0.0,0.0134017
0.308809,39.6742,0.0,0.0720468
2.32499,75.9855,0.0,0.0216741
0.966617,42.4246,0.0,0.0488755
5.83731,79.663,0.0,-0.03668019
2.316,38.0641,0.0,0.0769706
4.66148,79.663,0.0,-0.0415408
1.75822,39.9608,0.0,0.00252170
|
| Attachment 3: original_best_curved.png
|  |
| Attachment 4: best_curved_quad_zero.png
|  |
| Attachment 5: best_curved_same_radii.png
|  |
|
|
174
|
Fri Jul 15 18:33:20 2022 |
Alex M | Run Details: 2022_07_15_Latest_Greatest | We have started a new run. Here is the directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_07_15_Latest_Greatest
This run is a substantial overhaul. It is a curved run using the latest parameters from Ryan's optimization loop. Here is a list of important changes:
- The newest version of AraSim has been implemented.
- The minimum length has been decreased to 10 cm per side.
- The GA is recording the parents and operators used to generate individuals.
- The number of individuals has been increased to 100 per generation.
- The number of neutrinos per individual has been increased to 600k.
This run may take longer than previous runs due to the increased number of individuals. There may need to be modifications made to resolve or work around AraSim errors that delay the loop (due to the auto-resubmit function). Find the details of the run attached. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_07_15_Latest_Greatest' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=100 ## number of individuals per generation; please keep this value below 99
Seeds=5 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=120000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=12 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=76 ## Number (not fraction!) of individuals formed through crossover
MUTATION=10 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
173
|
Fri Jul 8 13:35:19 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | The second-best individual in this evolution was Bicone 22 from generation 40. This individual is, in fact, a fascinating case as we shall see. But to start the story of this individual we will go back to generation 38 in order to demonstrate some of the peculiarities.
In generation 38 there were two bicones of no renown, Bicone 11 and Bicone 17. Bicone 11 was a fairly average individual that was ranked 23rd with a fitness score of 4.72016. Its DNA was {*6.16084, ***79.663, 0.0015434, -0.107765} for side one , and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. One day, by chance it met Bicone 17, another average bicone ranked 24th with a score of 4.71877 and DNA {**2.32499, 79.663, -0.00224948, 0.192602} for side one, and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. These two individuals eventually became the parents of two antennas: Bicone 16 and Bicone 17 of generation 39.
Bicone 16 eventually grew to have been ranked 3rd in fitness score of 4.95323. It’s DNA ended up being a complete balance of its parents sharing side one with Bicone 38.11{2.32499, 79.663, -0.00224948, 0.192602} and side two with bicone 38.17 {0.308809, 39.6742, -0.0084247, 0.40629}. Bicone 16 was an individual with high aspirations and hoped to be reproduced. But alas, it was not meant to be. But bicone 16 came upon some amazing luck, it was selected with itself for crossover. This meant that bicone 16 was able to provide two identical surrogates to survive into the next generation. This is where this Bicone fulfilled its full potential.
The twin bicones were named Bicone 22 and Bicone 23 in generation 40. Being clones, they shared all their DNA with Bicone 39.16. However, due to some circumstances, they had slightly different fitness scores. Bicone 23 managed a very respectable 5.0117 fitness score and was ranked 2nd in the generation. But not to be outdone Bicone 22 managed to score a 5.17014 and ended up being the second-best performing individual of all time. Being so successful, the two bicones ended up producing 8 children between the two groups.
Bicone 23 was the first to crossover and had 2 children with its partner Bicone 25 (4.64409). These were Bicones 4 and 5. Bicone 4 was ranked 39th with a fitness score of 4.60251 and still shared the DNA of its second side with its grandparent Bicone 38.17 as well as most of its first side with Bicone 38.11 {2.32499, 79.663, -0.00213879, 0.192602} {0.308809, 39.9608, -0.0084247, 0.40629}. Bicone 5 on the other hand, was ranked 14th with a fitness score of 4.80535 and it still shared a lot of DNA with its grandparents {6.16084,53.0851,-0.00224948,0.0534469} {0.966617,39.6742,-0.0084247,0.40629}.
Bicone 22 had 6 children of its own with various other Bicones. These were; With Bicone 8.40 (4.82423) Bicone 8, ranked 11th with a fitness Score of 4.81784 and DNA {2.32499, 75.9855, -0.00224948, 0.192602} {0.966617, 39.6742, -0.00320023, 0.213833}; Bicone 9, ranked 7th with a fitness score of 4.88966 and DNA {6.10508, 79.663, -0.000594616, 0.0351901} {0.308809, 42.4246, -0.0084247, 0.40629}; With Biocone 16.40 (5.01137) Bicone 12, ranked 12th with a fitness score of 4.81705 and DNA {6.42695, 75.9855, 0.0015434, -0.107765} {0.308809, 39.6742, -0.00320023, 0.213833}; Bicone 13, ranked 2nd with a fitness score of 5.0344 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.966617, 42.4246, -0.0084247, 0.40629}; With Bicone 4.40 (4.66578) Bicone 34 ranked 3rd with a fitness score of 4.99864 and DNA {0.66148, 73.5522, -0.000594616, 0.00582814} {0.966617, 39.6742, -0.0084247, 0.40629}; and finally Bicone 35, ranked 22nd with fitness score 4.74955 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.308809, 42.4246, -0.0084247, 0.40629}
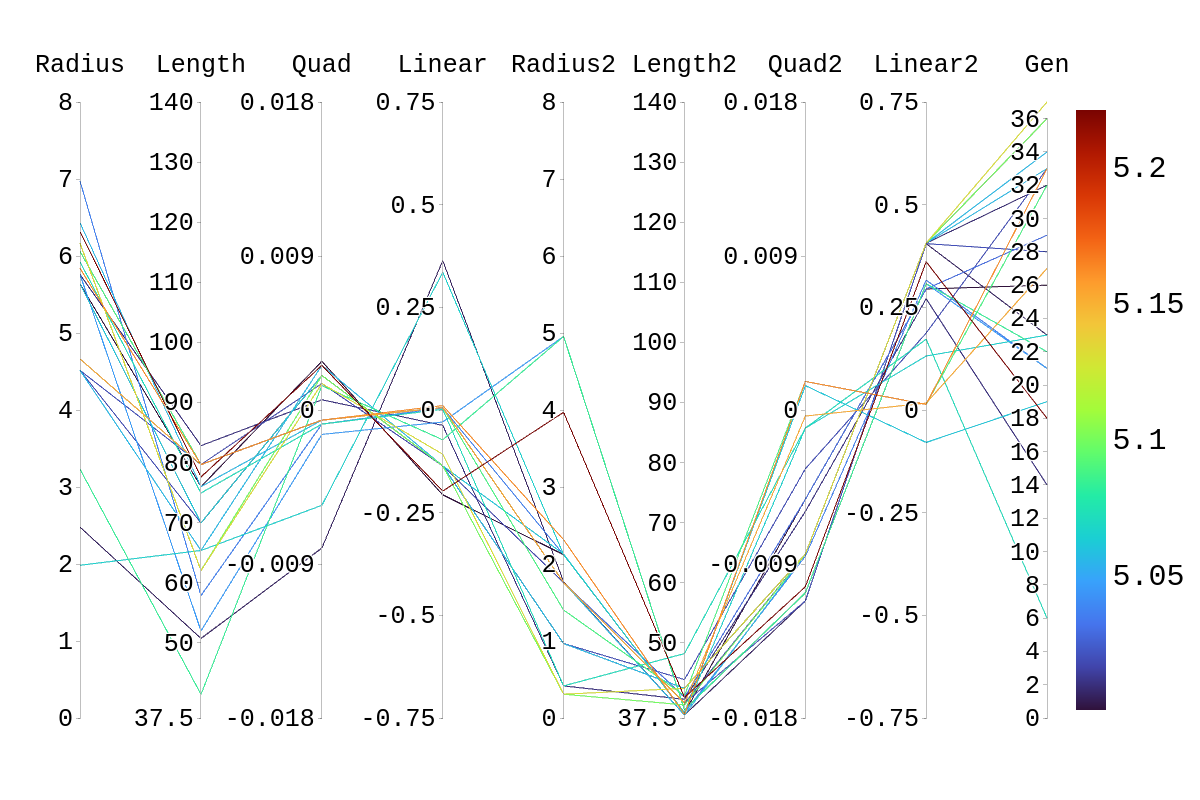
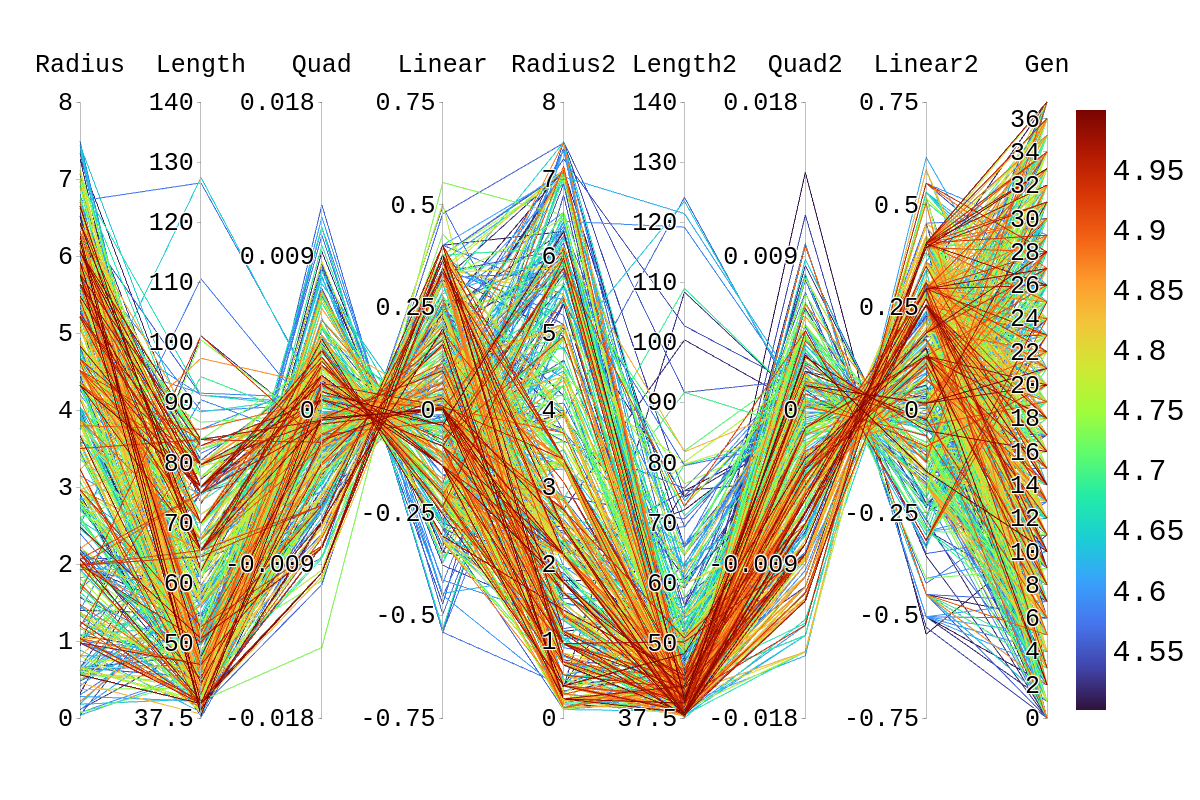
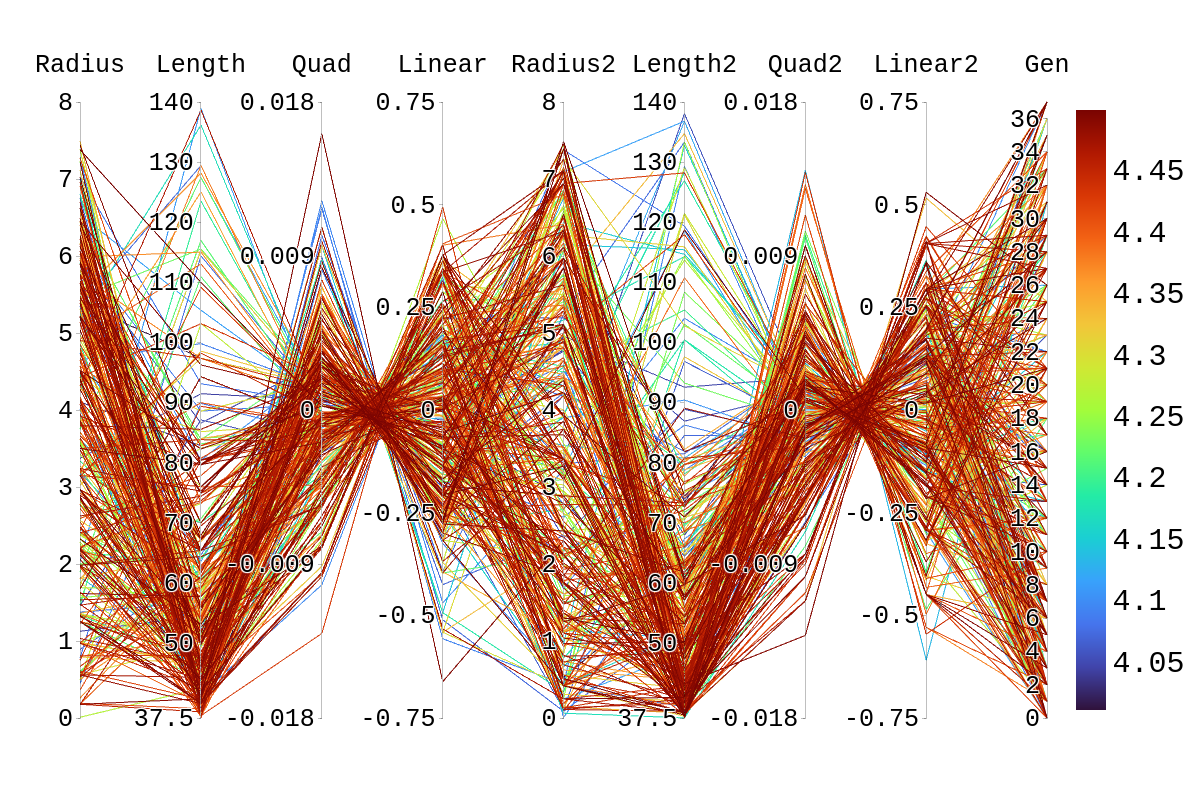
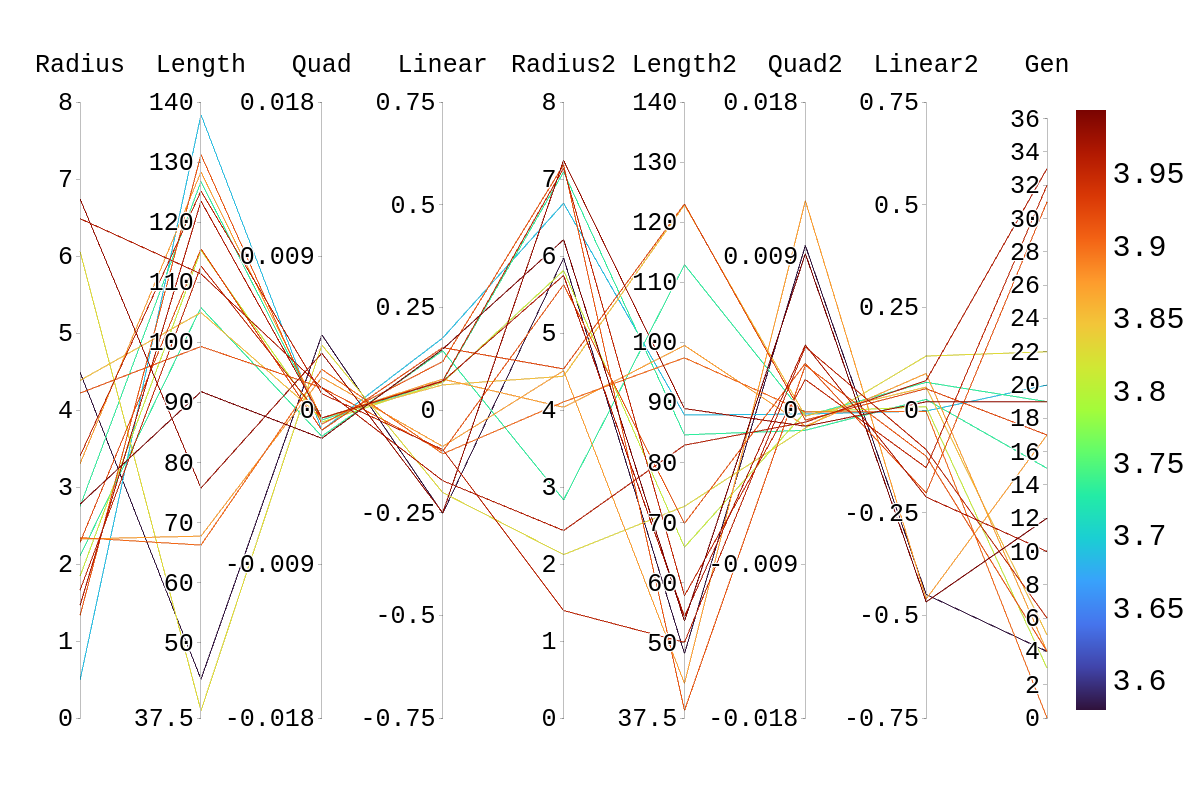
Upon inspection of the children in generation 41, you will see that none of the children surpassed their parent individual with the highest fitness score. They do however tend to have fitness scores around or above their second parent's score. Therefore, it is likely that the children's lower fitness score is due to a mismatch of genes. If we look at the rainbow plots provided, we can see that the genes of the worst-performing child are not dissimilar to those of individuals that scored higher than 5. This would imply that the fitness score found could be due to variations in Arasim similar to those seen from individuals 16.39 to 22/23.40 despite them being identical. Though it also could be possible that the graphs do not have a fine enough display to be able to clearly tell some of the genes apart.
*Gene originating from Bicone 38.11
**Gene originating from Bicone 38.17
***Gene originating from Bicone 38.11 and 38.17 that is shared between the two.
Amy: We want to nail things down in places where we still have hypotheses for what could have happened. For example, "This would imply that the fitness score found could be due to variations in Arasim similar to those seen from individuals 16.39 to 22/23.40 despite them being identical. Though it also could be possible that the graphs do not have a fine enough display to be able to clearly tell some of the genes apart." These two hypotheses can easily be distinguished by running with higher statistics and looking at the numbers rather than relying on the resolution of the display.
Amy: Also, at each stage, I was looking for the random numbers that were generated that led to the selection of the fractions of the population that were used for each type of selection and operator.
|
| Attachment 1: Rainbow_Plot_Quadratic_Rank_Test_5.png
|  |
| Attachment 2: Rainbow_Plot_Quadratic_Rank_Test_4.5.png
|  |
| Attachment 3: Rainbow_Plot_Quadratic_Rank_Test_4.png
|  |
| Attachment 4: Rainbow_Plot_Quadratic_Rank_Test_3.5.png
|  |
|
|
Draft
|
Fri Jul 8 13:34:08 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | |
|
|
Draft
|
Thu Jun 30 13:04:48 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | This is a multigenerational tracing of our second-best individual's parents and children:
The second-best individual in this evolution was Bicone 22 from generation 40. This individual is, in fact, a fascinating case as we shall see. But to start the story of this individual we will go back to generation 38 in order to demonstrate some of the peculiarities.
In generation 38 there were two bicones of no renown, Bicone 11 and Bicone 17. Bicone 11 was a fairly average individual that was ranked 23rd with a fitness score of 4.72016. Its DNA was {*6.16084, ***79.663, 0.0015434, -0.107765} for side one , and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. One day, by chance it met Bicone 17, another average bicone ranked 24th with a score of 4.71877 and DNA {**2.32499, 79.663, -0.00224948, 0.192602} for side one, and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. These two individuals eventually became the parents of two antennas: Bicone 16 and Bicone 17 of generation 39.
Bicone 16 eventually grew to have been ranked 3rd in fitness score of 4.95323. It’s DNA ended up being a complete balance of its parents sharing side one with Bicone 38.11{2.32499, 79.663, -0.00224948, 0.192602} and side two with bicone 38.17 {0.308809, 39.6742, -0.0084247, 0.40629}. Bicone 16 was an individual with high aspirations and hoped to be reproduced. But alas, it was not meant to be. But bicone 16 came upon some amazing luck, it was selected with itself for crossover. This meant that bicone 16 was able to provide two identical surrogates to survive into the next generation. This is where this Bicone fulfilled its full potential.
The twin bicones were named Bicone 22 and Bicone 23 in generation 40. Being clones, they shared all their DNA with Bicone 39.16. However, due to some circumstances, they had slightly different fitness scores. Bicone 23 managed a very respectable 5.0117 fitness score and was ranked 2nd in the generation. But not to be outdone Bicone 22 managed to score a 5.17014 and ended up being the second-best performing individual of all time. Being so successful, the two bicones ended up producing 8 children between the two groups.
Bicone 23 was the first to crossover and had 2 children with its partner. These were Bicones 4 and 5. Bicone 4 was ranked 39th with a fitness score of 4.60251 and still shared the DNA of its second side with its grandparent Bicone 38.17 as well as most of its first side with Bicone 38.11 {2.32499, 79.663, -0.00213879, 0.192602} {0.308809, 39.9608, -0.0084247, 0.40629}. Bicone 5 on the other hand, was ranked 14th with a fitness score of 4.80535 and it still shared a lot of DNA with its grandparents {6.16084,53.0851,-0.00224948,0.0534469} {0.966617,39.6742,-0.0084247,0.40629}.
Bicone 22 had 6 children of its own with various other Bicones. These were; Bicone 8, ranked 11th with a fitness Score of 4.81784 and DNA {2.32499, 75.9855, -0.00224948, 0.192602} {0.966617, 39.6742, -0.00320023, 0.213833}; Bicone 9, ranked 7th with a fitness score of 4.88966 and DNA {6.10508, 79.663, -0.000594616, 0.0351901} {0.308809, 42.4246, -0.0084247, 0.40629}; Bicone 12, ranked 12th with a fitness score of 4.81705 and DNA {6.42695, 75.9855, 0.0015434, -0.107765} {0.308809, 39.6742, -0.00320023, 0.213833}; Bicone 13, ranked 2nd with a fitness score of 5.0344 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.966617, 42.4246, -0.0084247, 0.40629}; Bicone 34 ranked 3rd with a fitness score of 4.99864 and DNA {0.66148, 73.5522, -0.000594616, 0.00582814} {0.966617, 39.6742, -0.0084247, 0.40629}; and finally Bicone 35, ranked 22nd with fitness score 4.74955 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.308809, 42.4246, -0.0084247, 0.40629}
Bellow, I have attached the rainbow plot with the parameters occupied by individual 4 in gen 41 which was again ranked 39th in that generation. From this, we can see that while in its own generation it was a poor performer, overall it was upper middle of the pack. However, because of the density of other better performing antennas in this region, it is hard to distinguish which genes in this antenna are contributing the most to the drop in fitness score compared to its siblings and parents.
*Gene originating from Bicone 38.11
**Gene originating from Bicone 38.17
***Gene originating from Bicone 38.11 and 38.17 that is shared between the two. |
| Attachment 1: rainbow_plot_quadratic_rank_test_720_(drawn).png
| .png) |
|
|
170
|
Tue Jun 28 13:27:13 2022 |
Dylan Wells | Changes needed for the matching circuit script |
-
Fix the functions for the SLPC, SCPL, and PLSC L networks (change the paramaters to match with the format of our data)
-
Write the PCSL function
-
Create a function to find the number of L networks necessary (N) given a source and load resistance as well as a frequency range.
-
Write a function to broadband match two impedances given a source, load, central frequency, and N. (return a list of capacitances and inductances for the L networks)
|
|
|
169
|
Tue Jun 28 12:49:53 2022 |
Dennis Calderon | Effective Volumes from AraSim: Curved Sides and Straight Sides (Paper Run) | Summary of results for 3million event simulations in AraSim with both GENETIS version and more recent {~11/2021) version.
Using errors for effective volume from the .out files.
Example shown below.
Radius: 3000 [m]
IceVolume : 8.4823e+10
test Veff(ice) : 6.50867e+09 m3sr, 6.50867 km3sr
test Veff(water eq.) : 5.96845e+09 m3sr, 5.96845 km3sr
And Veff(water eq.) error plus : 0.543588 and error minus : 0.543588
|
| Attachment 1: AraSim_Summary_Effective_Volumes.pdf
|
|
|
168
|
Tue Jun 28 11:40:32 2022 |
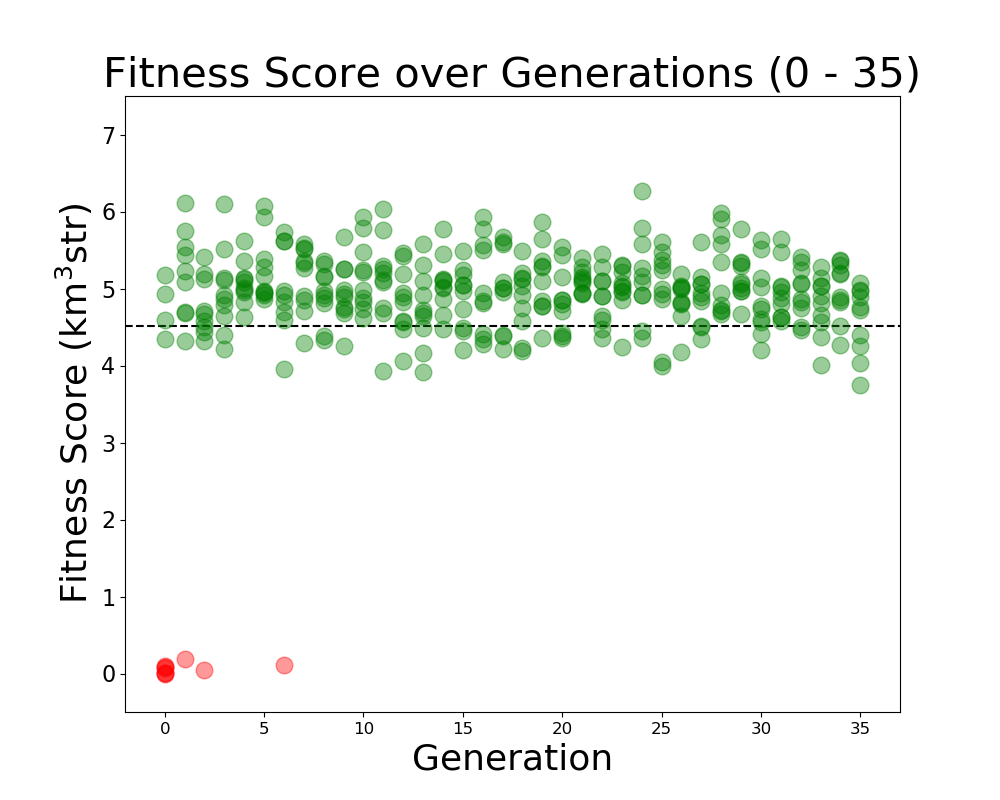
Alex M | Run Details: Machtay_20200911_Symmetric | This was a symmetric run that began on 9/11/2020. This run used fewer neutrinos to determine how significant the number would be on the performance of the evolution. 30,000 neutrinos were used per individual.
10 individuals were evolved over 35 generations, using 100% roulette selection and the old mutation algorithm, where genes had a 40% chance of mutating. Mutations changed the gene by reselecting the value from a guassian with a mean and sigma equalt to the mean and sigma of that gene's value in the generation.
|
| Attachment 1: Fitness_Scores_RG.png
|  |
|
|
167
|
Tue Jun 28 11:35:15 2022 |
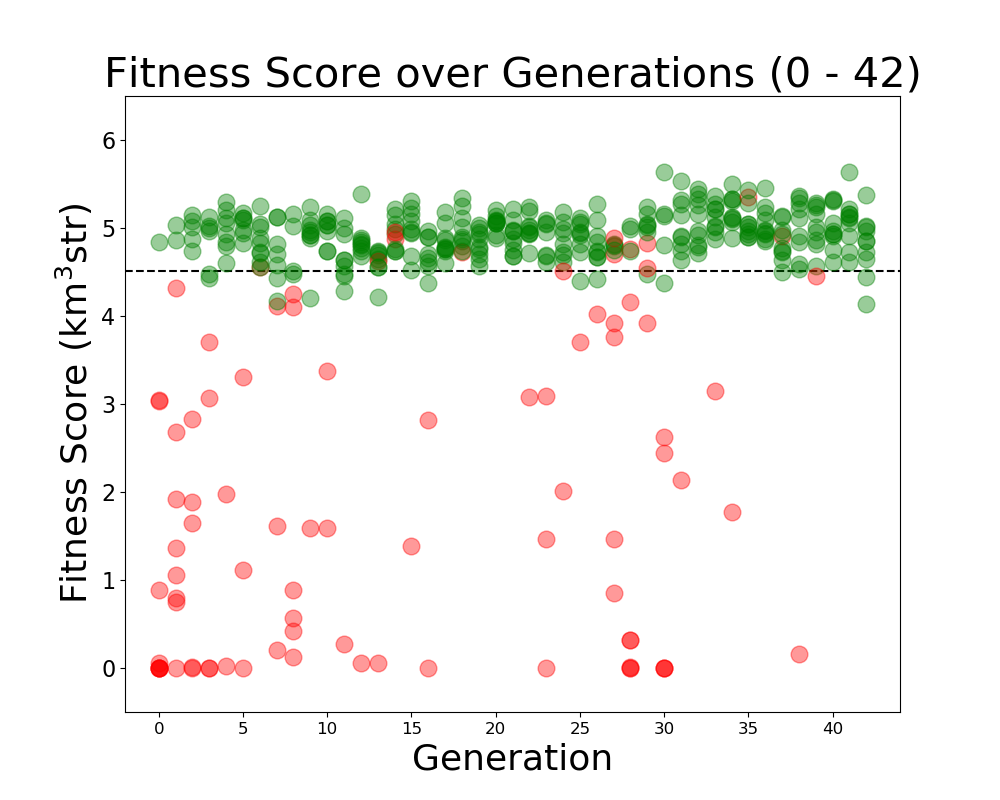
Alex M | Run Details: Machtay_20200831_Asym_Length_and_Angle | This run was conducted beginning on August 31, 2020.
Both the angle and length were asymmetric for the two cones. It was run with 10 individuals over 42 generations using 150,000 neutrinos. A penalty was still implemented on bicones exceeding the borehole width.
As this was before the substantial rewrite of the genetic algorithm, 100% of selection was done using roulette. Genes had a 40% chance to "mutate," which was done by selecting a new value from a guassian, with a mean and sigma equal to the mean and standard deviation of that gene's value in that generation.
|
| Attachment 1: Fitness_Scores_RG.png
|  |
|
|
166
|
Tue Jun 21 13:20:41 2022 |
Ryan Debolt | Multigenerational Narrative draft | The story of individual 8 from generation 18. (draft)
Once, there was a curved bicone named individual 8 who was from generation 18. In many ways, it was similar to many other bicones but in fact, this individual was the best bicone that ever lived with a fitness score of 5.22097. It achieved this by having the following parameters; an inner radius of 6.31483, and 3.97024; lengths of 77.6017 and 40.9244; quadratic coefficients of, 0.00260615 and, -0.0103313; and finally, a linear coefficient of -0.197494 and
0.36119. Unfortunately, individual 8’s lineage ended with it as it never met that special someone (crossover) and didn't survive into the next generation (reproduction) despite having a 77.7% of doing at least one of these.
This bicone had a sibling with a fitness Score of 4.65861 and had the following parameters: 6.05614,79.663,-0.000353038,0.0331969 (side 1)
1.92878,37.9849,-0.00213265,0.156724 (side 2).
Unfortunately, this bicone also suffered the same fate as its sibling and failed to leave a lasting mark on this hypothetical world.
These two individuals had parents from generation 17 whose name’s were individuals 39 and individual 8. After having individuals 8 and 9 in generation 18, individuals 39 and 8 from generation had a nasty divorce (possibly leading to 8 and 9’s aversion to marriage) and remarried. In these marriages, they both produced two more children that would be our previous antenna’s step-siblings.
Antenna 8 from gen 17 married antenna 46 to produce antenna 20 with a fitness score of 4.57122, and antenna 21 with a fitness score of 4.71056. Individual 20 shared the same genes for the second set of linear and quadratic coefficients as its more successful step-sibling individual 8. 21 had no similarities to individual 8 but had 4 children in generation 19.
Antenna 39 from gen 17 married antenna 5 to produce antenna 28 with a fitness score of 4.71808 and antenna 29 with a fitness score of 4.90119. Antenna 28 shared had an identical side 1 to individual 8 and also shared the same inner radius and length on the second side, and had 2 children in generation 19. Individual 29 had no similarities to individual 8 but really got around and had 8 children with various partners in generation 19. |
|
|
165
|
Tue Jun 21 12:22:02 2022 |
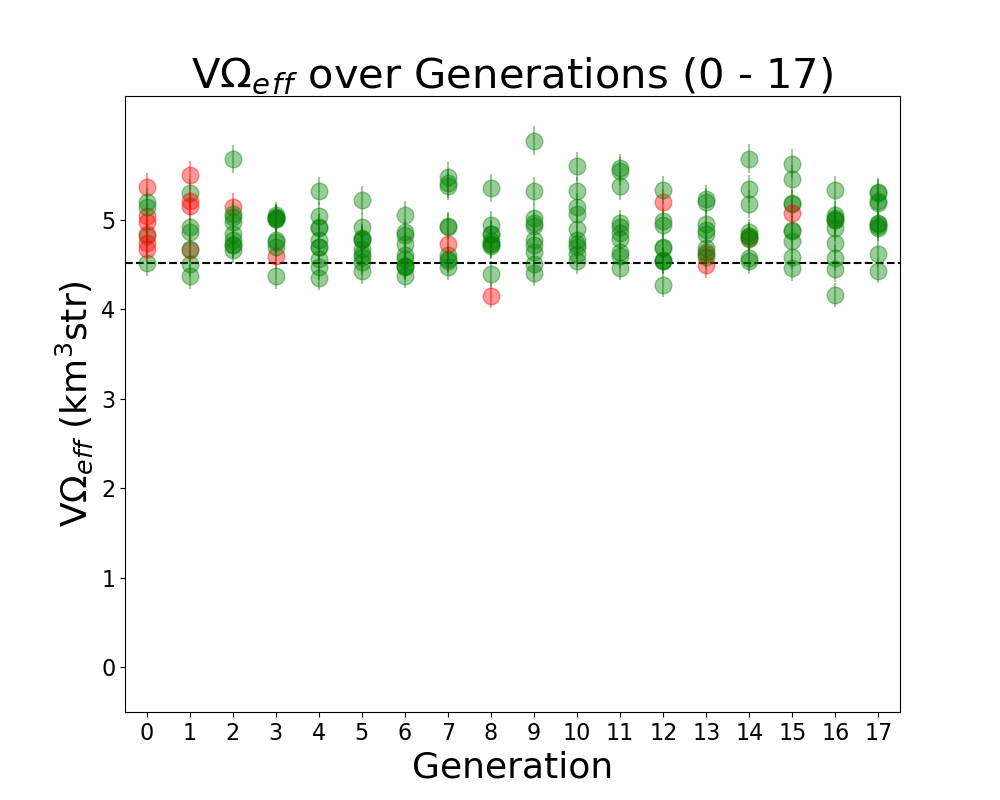
Alex M | Run Details: Machtay_20200827_Asym_Length_Run | This run followed the first symmetric run. It had an asymmetric length, but the opening angle and inner radius were kept symmetric.
10 individuals were evolved over 17 generations using 100,000 neutrinos. All selection was done through roulette, and all individuals were formed through crossover and the old mutation method (where individual genes had a 40% to be mutated, and mutated genes were chosen from a gaussian based on the mean and standard deviation of that gene's value in the generation). |
| Attachment 1: Veffectives_RG.png
|  |
| Attachment 2: Fitness_Scores_RG.png
| 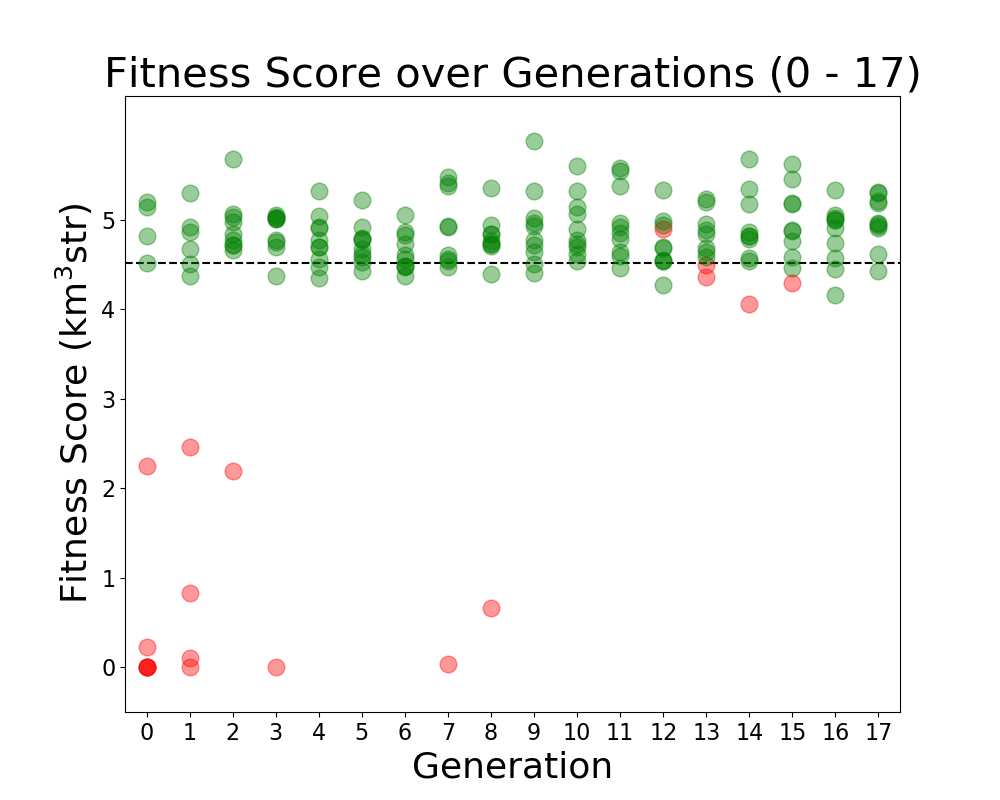 |
|
|
164
|
Tue Jun 21 12:15:18 2022 |
Alex M | Run Details: Machtay_20200824_Real_Run | This is considered to be the first run of "good" data. Prior to this run, we ran with few neutrinos and had errors which needed to be resolved in AraSim and with the way we handled XFdtd's simulation results being passed to AraSim. Those were resolved in the summer before this run.
This was a symmetric run of 10 individuals over 15 generations using 100,000 neutrinos per individual. This preceded the substantial modifications made by Ryan to the GA. All individuals were formed through crossover, but there was a probability of 40% for genes to be mutated. In this case, mutation was a complete change to the gene, though it was based on the average value of that gene in the generation. There was no reproduction or mutation. All individuals were selected through roulette.
Green individuals indicate that there was no penalty applied, while red individuals had a penalty factor multiplied by their effective volumes. The penalty was of the form exp(-(R-7.5)^2), where R is the outer radius, in cm, of the bicone. |
| Attachment 1: Fitness_Scores_RG.png
| 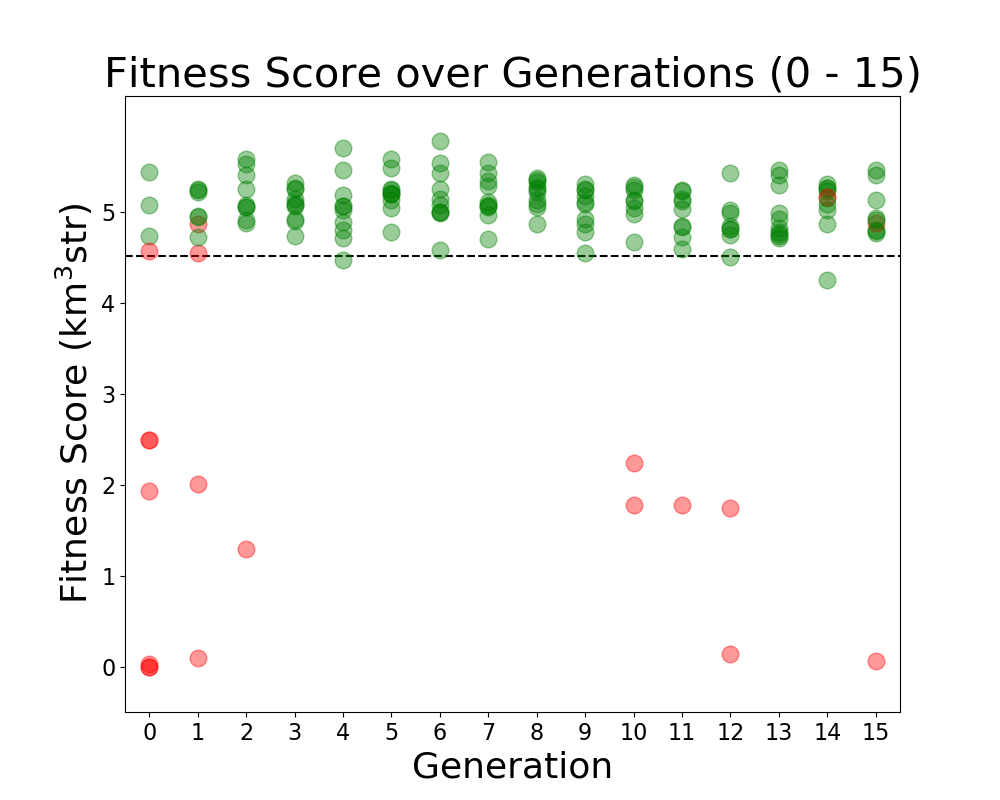 |
| Attachment 2: Veffectives_RG.png
| 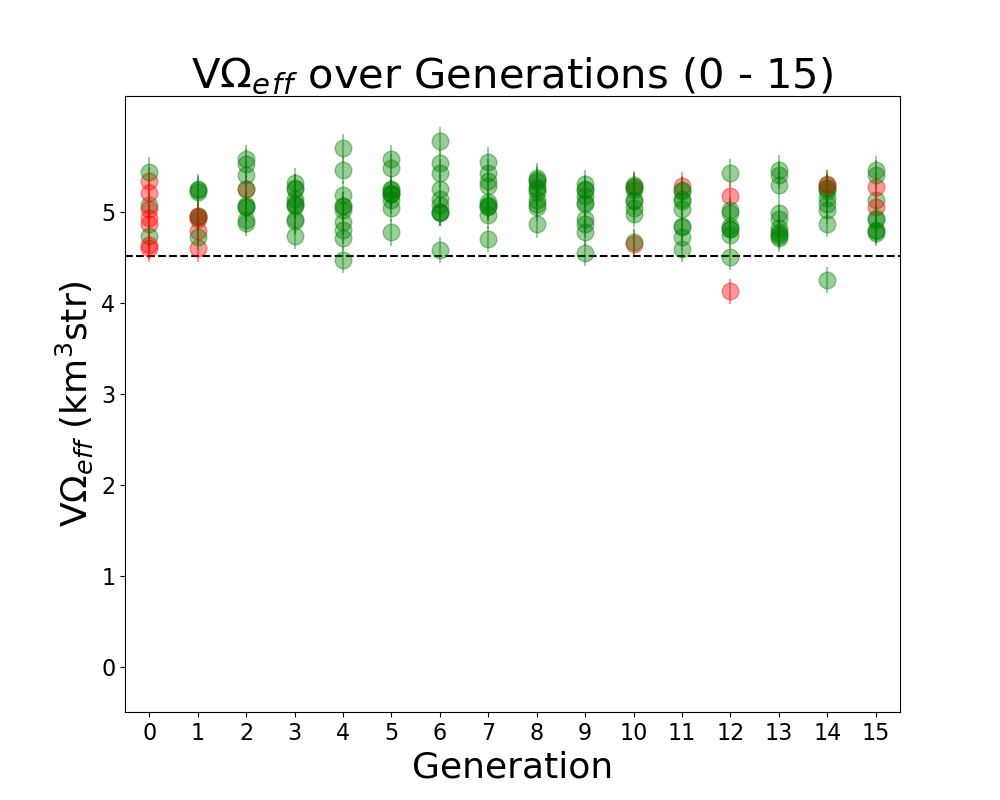 |
|