| ID |
Date |
Author |
Subject |
|
211
|
Tue Apr 11 00:36:49 2023 |
Bryan Reynolds | AREA physics of results plot first look |
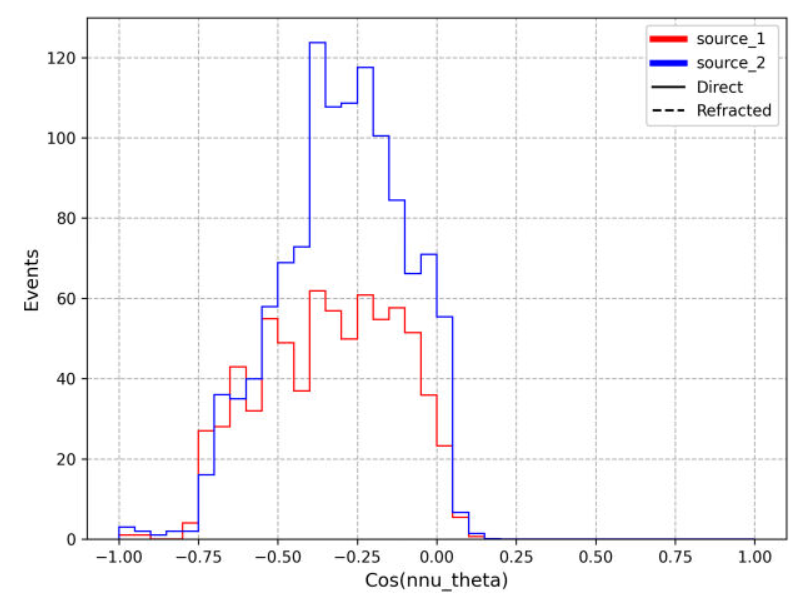
After some work, I've gotten the physics of results plotting script provided by Dennis working to compare the ARASim outputs from an AREA run to a set of reference data for the ARA bicone (provided by Alex). Among the outputs is a histogram of of events triggered vs cos(theta_nu) (attached here). This is the plot that was previously presented in GENETIS talks in the physics of results discussion.
Here I compared the best individual from the AREA results (Blue- "source_2") that I have on hand (only the ARASim output files from the most recent generation ran are currently saved) with the ARA bicone reference data (Red- "source_1"). Both datasets included a total of 150,000 NNU. Note that the AREA run used was a single frequency test, meaning that the radiation pattern was held constant for all frequencies in the band.
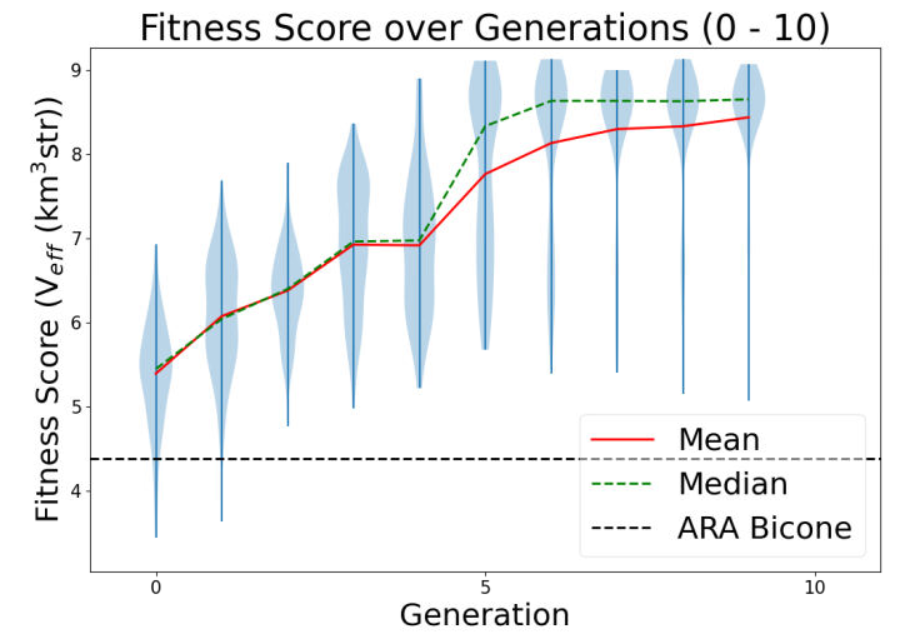
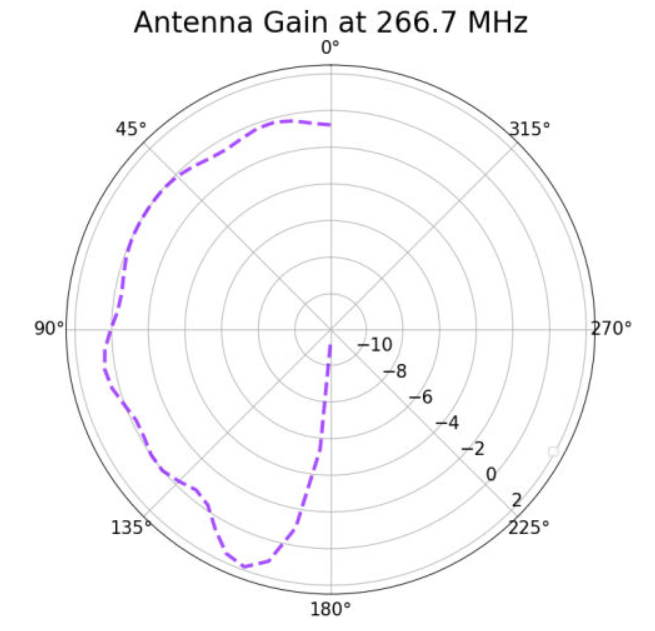
Also attached for reference are the gain pattern of the associated individual from the AREA output and the violin plot of the run producing the data. |
| Attachment 1: 20230410_AREA_PoR_CosTheta_bestIndiv_singleFreqTest.PNG
|
 |
| Attachment 2: 20230201_singleFreqTest_100NPop_fitnessPlot_10Gens.PNG
|
 |
| Attachment 3: 20230410_AREA_mostFitIndivReUpload_singleFreqTest.PNG
|
 |
|
189
|
Tue Jan 31 11:30:54 2023 |
Audrey Zinn | Building |
Attached are the XF files for the top 5 individuals from the 2022_12_29 crazy sides run. In order from best to 5th best:
1: Gen 29 Indiv 87
2: Gen 19 Indiv 96
3: Gen 13 Indiv 84
4: Gen 9 Indiv 50
5: Gen 18 Indiv 89
These can also be found in /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_12_29/2022_12_29.xf in the corresponding directories. |
| Attachment 1: XF_model_13_84.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -64.90 0.00 -105.60
5 0 -19.63 0.01 -105.58
10 0 -13.64 0.04 -105.51
15 0 -10.17 0.10 -105.38
20 0 -7.75 0.17 -105.21
25 0 -5.90 0.26 -104.96
30 0 -4.43 0.36 -104.65
35 0 -3.23 0.48 -104.25
40 0 -2.22 0.60 -103.75
45 0 -1.38 0.73 -103.11
50 0 -0.67 0.86 -102.31
55 0 -0.08 0.98 -101.26
60 0 0.42 1.10 -99.87
65 0 0.82 1.21 -97.94
70 0 1.14 1.30 -95.09
75 0 1.39 1.38 -90.53
80 0 1.56 1.43 -82.17
85 0 1.67 1.47 -63.52
90 0 1.70 1.48 -19.53
95 0 1.67 1.47 24.56
100 0 1.56 1.43 43.26
105 0 1.39 1.38 51.64
110 0 1.14 1.30 56.22
115 0 0.82 1.21 59.07
120 0 0.41 1.10 61.01
125 0 -0.08 0.98 62.41
130 0 -0.68 0.86 63.46
135 0 -1.39 0.73 64.28
140 0 -2.23 0.60 64.92
145 0 -3.23 0.48 65.43
150 0 -4.44 0.36 65.84
155 0 -5.91 0.26 66.17
160 0 -7.76 0.17 66.42
165 0 -10.19 0.10 66.62
170 0 -13.66 0.04 66.75
175 0 -19.66 0.01 66.84
180 0 -64.87 0.00 66.87
0 5 -64.33 0.00 -107.33
5 5 -19.63 0.01 -107.32
10 5 -13.64 0.04 -107.27
15 5 -10.17 0.10 -107.18
20 5 -7.75 0.17 -107.05
25 5 -5.90 0.26 -106.88
30 5 -4.43 0.36 -106.65
35 5 -3.23 0.48 -106.37
40 5 -2.22 0.60 -106.01
45 5 -1.38 0.73 -105.57
50 5 -0.67 0.86 -105.00
55 5 -0.08 0.98 -104.28
60 5 0.42 1.10 -103.31
65 5 0.82 1.21 -101.97
70 5 1.14 1.30 -99.99
75 5 1.39 1.38 -96.77
80 5 1.56 1.43 -90.60
85 5 1.67 1.47 -74.77
90 5 1.70 1.48 -18.14
95 5 1.67 1.47 36.69
100 5 1.56 1.43 51.96
105 5 1.39 1.38 57.99
110 5 1.14 1.30 61.17
115 5 0.82 1.21 63.13
120 5 0.41 1.10 64.46
125 5 -0.08 0.98 65.43
130 5 -0.68 0.86 66.16
135 5 -1.39 0.73 66.73
140 5 -2.23 0.60 67.19
145 5 -3.23 0.48 67.56
150 5 -4.44 0.36 67.85
155 5 -5.91 0.26 68.09
160 5 -7.76 0.17 68.27
165 5 -10.19 0.10 68.42
170 5 -13.66 0.04 68.52
175 5 -19.66 0.01 68.58
180 5 -64.30 0.00 68.61
0 10 -63.85 0.00 -109.51
5 10 -19.63 0.01 -109.51
10 10 -13.64 0.04 -109.49
15 10 -10.17 0.10 -109.45
20 10 -7.75 0.17 -109.38
25 10 -5.90 0.26 -109.30
30 10 -4.43 0.36 -109.19
35 10 -3.23 0.48 -109.06
40 10 -2.22 0.60 -108.89
45 10 -1.38 0.73 -108.69
50 10 -0.67 0.86 -108.44
55 10 -0.08 0.98 -108.13
60 10 0.42 1.10 -107.73
65 10 0.82 1.21 -107.19
70 10 1.14 1.30 -106.41
75 10 1.39 1.38 -105.17
80 10 1.56 1.43 -102.75
85 10 1.67 1.47 -95.68
90 10 1.70 1.48 -10.72
95 10 1.67 1.47 57.68
100 10 1.56 1.43 64.05
105 10 1.39 1.38 66.34
110 10 1.14 1.30 67.55
115 10 0.82 1.21 68.31
120 10 0.41 1.10 68.85
125 10 -0.08 0.98 69.26
130 10 -0.68 0.86 69.58
135 10 -1.39 0.73 69.84
140 10 -2.23 0.60 70.06
145 10 -3.23 0.48 70.24
150 10 -4.44 0.36 70.39
155 10 -5.91 0.26 70.51
160 10 -7.76 0.17 70.61
165 10 -10.19 0.10 70.69
170 10 -13.66 0.04 70.75
175 10 -19.66 0.01 70.78
180 10 -63.82 0.00 70.80
0 15 -63.45 0.00 -112.40
5 15 -19.63 0.01 -112.41
10 15 -13.64 0.04 -112.42
15 15 -10.17 0.10 -112.44
20 15 -7.75 0.17 -112.47
25 15 -5.90 0.26 -112.50
30 15 -4.43 0.36 -112.55
35 15 -3.23 0.48 -112.61
40 15 -2.22 0.60 -112.70
45 15 -1.38 0.73 -112.83
50 15 -0.67 0.86 -113.01
55 15 -0.08 0.98 -113.26
60 15 0.42 1.10 -113.63
65 15 0.82 1.21 -114.19
70 15 1.14 1.30 -115.09
75 15 1.39 1.38 -116.67
80 15 1.56 1.43 -119.98
85 15 1.67 1.47 -130.06
90 15 1.70 1.48 150.28
95 15 1.67 1.47 88.84
100 15 1.56 1.43 80.40
105 15 1.39 1.38 77.44
110 15 1.14 1.30 75.99
115 15 0.82 1.21 75.16
120 15 0.41 1.10 74.65
125 15 -0.08 0.98 74.32
130 15 -0.68 0.86 74.09
135 15 -1.39 0.73 73.95
140 15 -2.23 0.60 73.85
145 15 -3.23 0.48 73.78
150 15 -4.44 0.36 73.74
155 15 -5.91 0.26 73.71
160 15 -7.76 0.17 73.70
165 15 -10.19 0.10 73.69
170 15 -13.66 0.04 73.69
175 15 -19.66 0.01 73.70
180 15 -63.42 0.00 73.71
0 20 -63.14 0.00 -116.43
5 20 -19.63 0.01 -116.46
10 20 -13.64 0.04 -116.53
15 20 -10.17 0.10 -116.63
20 20 -7.75 0.17 -116.78
25 20 -5.90 0.26 -116.98
30 20 -4.43 0.36 -117.25
35 20 -3.23 0.48 -117.59
40 20 -2.22 0.60 -118.04
45 20 -1.38 0.73 -118.62
50 20 -0.67 0.86 -119.39
55 20 -0.08 0.98 -120.42
60 20 0.42 1.10 -121.84
65 20 0.82 1.21 -123.88
70 20 1.14 1.30 -126.97
75 20 1.39 1.38 -132.07
80 20 1.56 1.43 -141.54
85 20 1.67 1.47 -162.14
90 20 1.70 1.48 155.74
95 20 1.67 1.47 117.62
100 20 1.56 1.43 100.14
105 20 1.39 1.38 91.86
110 20 1.14 1.30 87.29
115 20 0.82 1.21 84.47
120 20 0.41 1.10 82.60
125 20 -0.08 0.98 81.29
130 20 -0.68 0.86 80.35
135 20 -1.39 0.73 79.64
140 20 -2.23 0.60 79.12
145 20 -3.23 0.48 78.72
150 20 -4.44 0.36 78.41
155 20 -5.91 0.26 78.18
160 20 -7.76 0.17 78.02
165 20 -10.19 0.10 77.90
170 20 -13.66 0.04 77.82
175 20 -19.66 0.01 77.78
180 20 -63.11 0.00 77.78
0 25 -62.89 0.00 -122.51
5 25 -19.63 0.01 -122.57
10 25 -13.64 0.04 -122.71
15 25 -10.17 0.10 -122.95
20 25 -7.75 0.17 -123.28
25 25 -5.90 0.26 -123.72
30 25 -4.43 0.36 -124.31
35 25 -3.23 0.48 -125.05
40 25 -2.22 0.60 -126.00
45 25 -1.38 0.73 -127.22
50 25 -0.67 0.86 -128.80
55 25 -0.08 0.98 -130.86
60 25 0.42 1.10 -133.62
65 25 0.82 1.21 -137.41
70 25 1.14 1.30 -142.79
75 25 1.39 1.38 -150.72
80 25 1.56 1.43 -162.72
85 25 1.67 1.47 179.55
90 25 1.70 1.48 157.02
95 25 1.67 1.47 135.52
100 25 1.56 1.43 119.63
105 25 1.39 1.38 109.06
110 25 1.14 1.30 102.05
115 25 0.82 1.21 97.25
120 25 0.41 1.10 93.83
125 25 -0.08 0.98 91.34
130 25 -0.68 0.86 89.46
135 25 -1.39 0.73 88.03
140 25 -2.23 0.60 86.93
145 25 -3.23 0.48 86.07
150 25 -4.44 0.36 85.40
155 25 -5.91 0.26 84.89
160 25 -7.76 0.17 84.50
165 25 -10.19 0.10 84.22
170 25 -13.66 0.04 84.04
175 25 -19.66 0.01 83.94
180 25 -62.87 0.00 83.92
0 30 -62.72 0.00 -132.56
5 30 -19.63 0.01 -132.68
10 30 -13.64 0.04 -132.94
15 30 -10.17 0.10 -133.37
20 30 -7.75 0.17 -133.97
25 30 -5.90 0.26 -134.77
30 30 -4.43 0.36 -135.79
35 30 -3.23 0.48 -137.09
40 30 -2.22 0.60 -138.71
45 30 -1.38 0.73 -140.72
50 30 -0.67 0.86 -143.24
55 30 -0.08 0.98 -146.39
60 30 0.42 1.10 -150.34
65 30 0.82 1.21 -155.35
70 30 1.14 1.30 -161.66
75 30 1.39 1.38 -169.57
80 30 1.56 1.43 -179.20
85 30 1.67 1.47 169.63
90 30 1.70 1.48 157.66
95 30 1.67 1.47 145.96
100 30 1.56 1.43 135.50
105 30 1.39 1.38 126.73
110 30 1.14 1.30 119.66
115 30 0.82 1.21 114.04
120 30 0.41 1.10 109.61
125 30 -0.08 0.98 106.09
130 30 -0.68 0.86 103.30
135 30 -1.39 0.73 101.07
140 30 -2.23 0.60 99.29
145 30 -3.23 0.48 97.86
150 30 -4.44 0.36 96.72
155 30 -5.91 0.26 95.83
160 30 -7.76 0.17 95.15
165 30 -10.19 0.10 94.66
170 30 -13.66 0.04 94.33
175 30 -19.66 0.01 94.15
180 30 -62.70 0.00 94.13
0 35 -62.62 0.00 -150.86
5 35 -19.63 0.01 -151.07
10 35 -13.64 0.04 -151.49
15 35 -10.17 0.10 -152.14
20 35 -7.75 0.17 -153.03
25 35 -5.90 0.26 -154.18
30 35 -4.43 0.36 -155.60
35 35 -3.23 0.48 -157.32
40 35 -2.22 0.60 -159.38
45 35 -1.38 0.73 -161.80
50 35 -0.67 0.86 -164.62
55 35 -0.08 0.98 -167.87
60 35 0.42 1.10 -171.57
65 35 0.82 1.21 -175.73
70 35 1.14 1.30 179.66
75 35 1.39 1.38 174.64
80 35 1.56 1.43 169.28
85 35 1.67 1.47 163.72
90 35 1.70 1.48 158.08
95 35 1.67 1.47 152.52
100 35 1.56 1.43 147.18
105 35 1.39 1.38 142.18
110 35 1.14 1.30 137.59
115 35 0.82 1.21 133.45
120 35 0.41 1.10 129.78
125 35 -0.08 0.98 126.55
130 35 -0.68 0.86 123.75
135 35 -1.39 0.73 121.34
140 35 -2.23 0.60 119.29
145 35 -3.23 0.48 117.57
150 35 -4.44 0.36 116.14
155 35 -5.91 0.26 114.99
160 35 -7.76 0.17 114.09
165 35 -10.19 0.10 113.43
170 35 -13.66 0.04 113.00
175 35 -19.66 0.01 112.78
180 35 -62.60 0.00 112.77
0 40 -62.58 0.00 177.48
... 159721 more lines ...
|
| Attachment 2: XF_model_9_50.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -65.66 0.00 68.97
5 0 -23.00 0.01 68.99
10 0 -17.02 0.02 69.05
15 0 -13.55 0.04 69.15
20 0 -11.13 0.08 69.30
25 0 -9.29 0.12 69.49
30 0 -7.82 0.17 69.75
35 0 -6.63 0.22 70.06
40 0 -5.63 0.27 70.43
45 0 -4.79 0.33 70.90
50 0 -4.09 0.39 71.47
55 0 -3.50 0.45 72.18
60 0 -3.01 0.50 73.09
65 0 -2.61 0.55 74.31
70 0 -2.29 0.59 76.06
75 0 -2.05 0.62 78.83
80 0 -1.88 0.65 84.06
85 0 -1.78 0.66 97.84
90 0 -1.74 0.67 159.48
95 0 -1.78 0.66 -139.66
100 0 -1.88 0.65 -126.04
105 0 -2.05 0.62 -120.85
110 0 -2.30 0.59 -118.08
115 0 -2.61 0.55 -116.34
120 0 -3.02 0.50 -115.12
125 0 -3.51 0.45 -114.20
130 0 -4.10 0.39 -113.48
135 0 -4.80 0.33 -112.91
140 0 -5.63 0.27 -112.44
145 0 -6.63 0.22 -112.05
150 0 -7.83 0.16 -111.73
155 0 -9.30 0.12 -111.47
160 0 -11.14 0.08 -111.26
165 0 -13.57 0.04 -111.10
170 0 -17.04 0.02 -110.98
175 0 -23.04 0.00 -110.91
180 0 -65.62 0.00 -110.88
0 5 -66.34 0.00 70.70
5 5 -23.00 0.01 70.72
10 5 -17.02 0.02 70.80
15 5 -13.55 0.04 70.93
20 5 -11.13 0.08 71.12
25 5 -9.29 0.12 71.39
30 5 -7.82 0.17 71.72
35 5 -6.63 0.22 72.14
40 5 -5.63 0.27 72.65
45 5 -4.79 0.33 73.29
50 5 -4.09 0.39 74.09
55 5 -3.50 0.45 75.11
60 5 -3.01 0.50 76.44
65 5 -2.61 0.55 78.24
70 5 -2.29 0.59 80.87
75 5 -2.05 0.62 85.05
80 5 -1.88 0.65 92.78
85 5 -1.78 0.66 110.94
90 5 -1.74 0.67 160.03
95 5 -1.78 0.66 -152.08
100 5 -1.88 0.65 -134.51
105 5 -2.05 0.62 -126.96
110 5 -2.30 0.59 -122.85
115 5 -2.61 0.55 -120.26
120 5 -3.02 0.50 -118.46
125 5 -3.51 0.45 -117.14
130 5 -4.10 0.39 -116.13
135 5 -4.80 0.33 -115.32
140 5 -5.63 0.27 -114.67
145 5 -6.63 0.22 -114.15
150 5 -7.83 0.16 -113.72
155 5 -9.30 0.12 -113.38
160 5 -11.14 0.08 -113.10
165 5 -13.57 0.04 -112.89
170 5 -17.04 0.02 -112.75
175 5 -23.04 0.00 -112.66
180 5 -66.29 0.00 -112.62
0 10 -67.14 0.00 72.17
5 10 -23.00 0.01 72.19
10 10 -17.02 0.02 72.28
15 10 -13.55 0.04 72.45
20 10 -11.13 0.08 72.68
25 10 -9.29 0.12 72.99
30 10 -7.82 0.17 73.40
35 10 -6.63 0.22 73.91
40 10 -5.63 0.27 74.54
45 10 -4.79 0.33 75.33
50 10 -4.09 0.39 76.32
55 10 -3.50 0.45 77.59
60 10 -3.01 0.50 79.26
65 10 -2.61 0.55 81.54
70 10 -2.29 0.59 84.86
75 10 -2.05 0.62 90.10
80 10 -1.88 0.65 99.46
85 10 -1.78 0.66 119.21
90 10 -1.74 0.67 160.25
95 10 -1.78 0.66 -159.82
100 10 -1.88 0.65 -140.91
105 10 -2.05 0.62 -131.87
110 10 -2.30 0.59 -126.77
115 10 -2.61 0.55 -123.52
120 10 -3.02 0.50 -121.27
125 10 -3.51 0.45 -119.61
130 10 -4.10 0.39 -118.35
135 10 -4.80 0.33 -117.36
140 10 -5.63 0.27 -116.57
145 10 -6.63 0.22 -115.93
150 10 -7.83 0.16 -115.41
155 10 -9.30 0.12 -114.99
160 10 -11.14 0.08 -114.67
165 10 -13.57 0.04 -114.42
170 10 -17.04 0.02 -114.25
175 10 -23.04 0.00 -114.14
180 10 -67.09 0.00 -114.10
0 15 -68.08 0.00 73.45
5 15 -23.00 0.01 73.48
10 15 -17.02 0.02 73.58
15 15 -13.55 0.04 73.77
20 15 -11.13 0.08 74.04
25 15 -9.29 0.12 74.40
30 15 -7.82 0.17 74.86
35 15 -6.63 0.22 75.45
40 15 -5.63 0.27 76.18
45 15 -4.79 0.33 77.10
50 15 -4.09 0.39 78.26
55 15 -3.50 0.45 79.74
60 15 -3.01 0.50 81.70
65 15 -2.61 0.55 84.37
70 15 -2.29 0.59 88.24
75 15 -2.05 0.62 94.27
80 15 -1.88 0.65 104.69
85 15 -1.78 0.66 124.77
90 15 -1.74 0.67 160.35
95 15 -1.78 0.66 -165.00
100 15 -1.88 0.65 -145.87
105 15 -2.05 0.62 -135.89
110 15 -2.30 0.59 -130.06
115 15 -2.61 0.55 -126.29
120 15 -3.02 0.50 -123.67
125 15 -3.51 0.45 -121.74
130 15 -4.10 0.39 -120.28
135 15 -4.80 0.33 -119.13
140 15 -5.63 0.27 -118.21
145 15 -6.63 0.22 -117.47
150 15 -7.83 0.16 -116.88
155 15 -9.30 0.12 -116.40
160 15 -11.14 0.08 -116.03
165 15 -13.57 0.04 -115.75
170 15 -17.04 0.02 -115.55
175 15 -23.04 0.00 -115.43
180 15 -68.02 0.00 -115.39
0 20 -69.19 0.00 74.59
5 20 -23.00 0.01 74.62
10 20 -17.02 0.02 74.74
15 20 -13.55 0.04 74.95
20 20 -11.13 0.08 75.25
25 20 -9.29 0.12 75.65
30 20 -7.82 0.17 76.17
35 20 -6.63 0.22 76.82
40 20 -5.63 0.27 77.64
45 20 -4.79 0.33 78.67
50 20 -4.09 0.39 79.97
55 20 -3.50 0.45 81.64
60 20 -3.01 0.50 83.84
65 20 -2.61 0.55 86.85
70 20 -2.29 0.59 91.16
75 20 -2.05 0.62 97.79
80 20 -1.88 0.65 108.88
85 20 -1.78 0.66 128.73
90 20 -1.74 0.67 160.41
95 20 -1.78 0.66 -168.71
100 20 -1.88 0.65 -149.83
105 20 -2.05 0.62 -139.26
110 20 -2.30 0.59 -132.89
115 20 -2.61 0.55 -128.71
120 20 -3.02 0.50 -125.78
125 20 -3.51 0.45 -123.62
130 20 -4.10 0.39 -121.98
135 20 -4.80 0.33 -120.69
140 20 -5.63 0.27 -119.66
145 20 -6.63 0.22 -118.84
150 20 -7.83 0.16 -118.18
155 20 -9.30 0.12 -117.66
160 20 -11.14 0.08 -117.24
165 20 -13.57 0.04 -116.93
170 20 -17.04 0.02 -116.72
175 20 -23.04 0.00 -116.58
180 20 -69.13 0.00 -116.53
0 25 -70.51 0.00 75.63
5 25 -23.00 0.01 75.67
10 25 -17.02 0.02 75.80
15 25 -13.56 0.04 76.02
20 25 -11.13 0.08 76.35
25 25 -9.29 0.12 76.79
30 25 -7.82 0.17 77.35
35 25 -6.63 0.22 78.07
40 25 -5.63 0.27 78.97
45 25 -4.79 0.33 80.10
50 25 -4.09 0.39 81.52
55 25 -3.50 0.45 83.36
60 25 -3.01 0.50 85.77
65 25 -2.61 0.55 89.05
70 25 -2.29 0.59 93.74
75 25 -2.05 0.62 100.82
80 25 -1.88 0.65 112.32
85 25 -1.78 0.66 131.71
90 25 -1.74 0.67 160.43
95 25 -1.78 0.66 -171.53
100 25 -1.88 0.65 -153.09
105 25 -2.05 0.62 -142.16
110 25 -2.30 0.59 -135.37
115 25 -2.61 0.55 -130.85
120 25 -3.02 0.50 -127.66
125 25 -3.51 0.45 -125.31
130 25 -4.10 0.39 -123.51
135 25 -4.80 0.33 -122.10
140 25 -5.63 0.27 -120.98
145 25 -6.63 0.22 -120.09
150 25 -7.83 0.16 -119.37
155 25 -9.30 0.12 -118.79
160 25 -11.14 0.08 -118.35
165 25 -13.57 0.04 -118.01
170 25 -17.04 0.02 -117.77
175 25 -23.04 0.00 -117.63
180 25 -70.44 0.00 -117.58
0 30 -72.08 0.00 76.59
5 30 -23.00 0.01 76.63
10 30 -17.02 0.02 76.77
15 30 -13.56 0.04 77.02
20 30 -11.13 0.08 77.37
25 30 -9.29 0.12 77.84
30 30 -7.82 0.17 78.45
35 30 -6.63 0.22 79.23
40 30 -5.63 0.27 80.20
45 30 -4.79 0.33 81.41
50 30 -4.09 0.39 82.96
55 30 -3.50 0.45 84.93
60 30 -3.01 0.50 87.53
65 30 -2.61 0.55 91.06
70 30 -2.29 0.59 96.04
75 30 -2.05 0.62 103.48
80 30 -1.88 0.65 115.21
85 30 -1.78 0.66 134.04
90 30 -1.74 0.67 160.43
95 30 -1.78 0.66 -173.76
100 30 -1.88 0.65 -155.84
105 30 -2.05 0.62 -144.70
110 30 -2.30 0.59 -137.59
115 30 -2.61 0.55 -132.79
120 30 -3.02 0.50 -129.38
125 30 -3.51 0.45 -126.85
130 30 -4.10 0.39 -124.91
135 30 -4.80 0.33 -123.40
140 30 -5.63 0.27 -122.20
145 30 -6.63 0.22 -121.23
150 30 -7.83 0.16 -120.46
155 30 -9.30 0.12 -119.84
160 30 -11.14 0.08 -119.36
165 30 -13.57 0.04 -119.00
170 30 -17.04 0.02 -118.75
175 30 -23.04 0.00 -118.60
180 30 -72.01 0.00 -118.54
0 35 -73.95 0.00 77.50
5 35 -23.00 0.01 77.55
10 35 -17.02 0.02 77.70
15 35 -13.56 0.04 77.95
20 35 -11.13 0.08 78.33
25 35 -9.29 0.12 78.83
30 35 -7.82 0.17 79.49
35 35 -6.63 0.22 80.31
40 35 -5.63 0.27 81.35
45 35 -4.79 0.33 82.65
50 35 -4.09 0.39 84.29
55 35 -3.50 0.45 86.40
60 35 -3.01 0.50 89.17
65 35 -2.61 0.55 92.90
70 35 -2.29 0.59 98.15
75 35 -2.05 0.62 105.86
80 35 -1.88 0.65 117.70
85 35 -1.78 0.66 135.93
90 35 -1.74 0.67 160.42
95 35 -1.78 0.66 -175.60
100 35 -1.88 0.65 -158.22
105 35 -2.05 0.62 -146.97
110 35 -2.30 0.59 -139.62
115 35 -2.61 0.55 -134.58
120 35 -3.02 0.50 -130.97
125 35 -3.51 0.45 -128.29
130 35 -4.10 0.39 -126.23
135 35 -4.80 0.33 -124.62
140 35 -5.63 0.27 -123.33
145 35 -6.63 0.22 -122.31
150 35 -7.83 0.16 -121.49
155 35 -9.30 0.12 -120.83
160 35 -11.14 0.08 -120.32
165 35 -13.57 0.04 -119.94
170 35 -17.04 0.02 -119.67
175 35 -23.04 0.00 -119.51
180 35 -73.87 0.00 -119.46
0 40 -76.10 0.00 78.37
... 159721 more lines ...
|
| Attachment 3: XF_model_18_89.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -300.00 0.00 -25.63
5 0 -300.00 0.00 -25.69
10 0 -300.00 0.00 -25.72
15 0 -300.00 0.00 -25.73
20 0 -300.00 0.00 -25.71
25 0 -300.00 0.00 -25.66
30 0 -300.00 0.00 -25.58
35 0 -300.00 0.00 -25.48
40 0 -300.00 0.00 -25.35
45 0 -300.00 0.00 -25.20
50 0 -300.00 0.00 -25.02
55 0 -300.00 0.00 -24.82
60 0 -300.00 0.00 -24.59
65 0 -300.00 0.00 -24.33
70 0 -300.00 0.00 -24.06
75 0 -300.00 0.00 -23.76
80 0 -300.00 0.00 -23.44
85 0 -300.00 0.00 -23.11
90 0 -300.00 0.00 -22.76
95 0 -300.00 0.00 -22.39
100 0 -300.00 0.00 -22.02
105 0 -300.00 0.00 -21.63
110 0 -300.00 0.00 -21.24
115 0 -300.00 0.00 -20.85
120 0 -300.00 0.00 -20.46
125 0 -300.00 0.00 -20.07
130 0 -300.00 0.00 -19.70
135 0 -300.00 0.00 -19.34
140 0 -300.00 0.00 -18.99
145 0 -300.00 0.00 -18.67
150 0 -300.00 0.00 -18.37
155 0 -300.00 0.00 -18.09
160 0 -300.00 0.00 -17.85
165 0 -300.00 0.00 -17.64
170 0 -300.00 0.00 -17.47
175 0 -300.00 0.00 -17.33
180 0 -300.00 0.00 -17.23
0 5 -300.00 0.00 -57.01
5 5 -300.00 0.00 -56.84
10 5 -300.00 0.00 -56.42
15 5 -300.00 0.00 -55.73
20 5 -300.00 0.00 -54.77
25 5 -300.00 0.00 -53.53
30 5 -300.00 0.00 -52.01
35 5 -300.00 0.00 -50.20
40 5 -300.00 0.00 -48.07
45 5 -300.00 0.00 -45.62
50 5 -300.00 0.00 -42.84
55 5 -300.00 0.00 -39.72
60 5 -300.00 0.00 -36.29
65 5 -300.00 0.00 -32.55
70 5 -300.00 0.00 -28.55
75 5 -300.00 0.00 -24.35
80 5 -300.00 0.00 -20.01
85 5 -300.00 0.00 -15.63
90 5 -300.00 0.00 -11.28
95 5 -300.00 0.00 -7.06
100 5 -300.00 0.00 -3.05
105 5 -300.00 0.00 0.71
110 5 -300.00 0.00 4.17
115 5 -300.00 0.00 7.31
120 5 -300.00 0.00 10.11
125 5 -300.00 0.00 12.60
130 5 -300.00 0.00 14.77
135 5 -300.00 0.00 16.65
140 5 -300.00 0.00 18.25
145 5 -300.00 0.00 19.59
150 5 -300.00 0.00 20.70
155 5 -300.00 0.00 21.58
160 5 -300.00 0.00 22.25
165 5 -300.00 0.00 22.74
170 5 -300.00 0.00 23.03
175 5 -300.00 0.00 23.15
180 5 -300.00 0.00 23.10
0 10 -300.00 0.00 -64.53
5 10 -300.00 0.00 -64.32
10 10 -300.00 0.00 -63.83
15 10 -300.00 0.00 -63.05
20 10 -300.00 0.00 -61.97
25 10 -300.00 0.00 -60.57
30 10 -300.00 0.00 -58.84
35 10 -300.00 0.00 -56.75
40 10 -300.00 0.00 -54.27
45 10 -300.00 0.00 -51.37
50 10 -300.00 0.00 -48.02
55 10 -300.00 0.00 -44.20
60 10 -300.00 0.00 -39.91
65 10 -300.00 0.00 -35.16
70 10 -300.00 0.00 -30.01
75 10 -300.00 0.00 -24.54
80 10 -300.00 0.00 -18.89
85 10 -300.00 0.00 -13.22
90 10 -300.00 0.00 -7.66
95 10 -300.00 0.00 -2.37
100 10 -300.00 0.00 2.54
105 10 -300.00 0.00 7.02
110 10 -300.00 0.00 11.02
115 10 -300.00 0.00 14.56
120 10 -300.00 0.00 17.64
125 10 -300.00 0.00 20.29
130 10 -300.00 0.00 22.57
135 10 -300.00 0.00 24.49
140 10 -300.00 0.00 26.11
145 10 -300.00 0.00 27.44
150 10 -300.00 0.00 28.52
155 10 -300.00 0.00 29.37
160 10 -300.00 0.00 30.01
165 10 -300.00 0.00 30.45
170 10 -300.00 0.00 30.71
175 10 -300.00 0.00 30.79
180 10 -300.00 0.00 30.71
0 15 -300.00 0.00 -67.78
5 15 -300.00 0.00 -67.56
10 15 -300.00 0.00 -67.05
15 15 -300.00 0.00 -66.24
20 15 -300.00 0.00 -65.13
25 15 -300.00 0.00 -63.69
30 15 -300.00 0.00 -61.90
35 15 -300.00 0.00 -59.72
40 15 -300.00 0.00 -57.12
45 15 -300.00 0.00 -54.05
50 15 -300.00 0.00 -50.47
55 15 -300.00 0.00 -46.35
60 15 -300.00 0.00 -41.67
65 15 -300.00 0.00 -36.45
70 15 -300.00 0.00 -30.73
75 15 -300.00 0.00 -24.64
80 15 -300.00 0.00 -18.34
85 15 -300.00 0.00 -12.03
90 15 -300.00 0.00 -5.90
95 15 -300.00 0.00 -0.13
100 15 -300.00 0.00 5.17
105 15 -300.00 0.00 9.92
110 15 -300.00 0.00 14.11
115 15 -300.00 0.00 17.75
120 15 -300.00 0.00 20.89
125 15 -300.00 0.00 23.58
130 15 -300.00 0.00 25.85
135 15 -300.00 0.00 27.76
140 15 -300.00 0.00 29.35
145 15 -300.00 0.00 30.65
150 15 -300.00 0.00 31.70
155 15 -300.00 0.00 32.52
160 15 -300.00 0.00 33.14
165 15 -300.00 0.00 33.56
170 15 -300.00 0.00 33.80
175 15 -300.00 0.00 33.87
180 15 -300.00 0.00 33.78
0 20 -300.00 0.00 -69.61
5 20 -300.00 0.00 -69.38
10 20 -300.00 0.00 -68.87
15 20 -300.00 0.00 -68.05
20 20 -300.00 0.00 -66.92
25 20 -300.00 0.00 -65.47
30 20 -300.00 0.00 -63.65
35 20 -300.00 0.00 -61.43
40 20 -300.00 0.00 -58.77
45 20 -300.00 0.00 -55.61
50 20 -300.00 0.00 -51.92
55 20 -300.00 0.00 -47.63
60 20 -300.00 0.00 -42.73
65 20 -300.00 0.00 -37.23
70 20 -300.00 0.00 -31.18
75 20 -300.00 0.00 -24.71
80 20 -300.00 0.00 -18.02
85 20 -300.00 0.00 -11.32
90 20 -300.00 0.00 -4.86
95 20 -300.00 0.00 1.18
100 20 -300.00 0.00 6.68
105 20 -300.00 0.00 11.57
110 20 -300.00 0.00 15.84
115 20 -300.00 0.00 19.54
120 20 -300.00 0.00 22.70
125 20 -300.00 0.00 25.38
130 20 -300.00 0.00 27.64
135 20 -300.00 0.00 29.54
140 20 -300.00 0.00 31.11
145 20 -300.00 0.00 32.39
150 20 -300.00 0.00 33.42
155 20 -300.00 0.00 34.23
160 20 -300.00 0.00 34.83
165 20 -300.00 0.00 35.24
170 20 -300.00 0.00 35.47
175 20 -300.00 0.00 35.54
180 20 -300.00 0.00 35.45
0 25 -300.00 0.00 -70.79
5 25 -300.00 0.00 -70.56
10 25 -300.00 0.00 -70.04
15 25 -300.00 0.00 -69.23
20 25 -300.00 0.00 -68.10
25 25 -300.00 0.00 -66.63
30 25 -300.00 0.00 -64.80
35 25 -300.00 0.00 -62.56
40 25 -300.00 0.00 -59.86
45 25 -300.00 0.00 -56.66
50 25 -300.00 0.00 -52.88
55 25 -300.00 0.00 -48.49
60 25 -300.00 0.00 -43.45
65 25 -300.00 0.00 -37.76
70 25 -300.00 0.00 -31.49
75 25 -300.00 0.00 -24.77
80 25 -300.00 0.00 -17.81
85 25 -300.00 0.00 -10.86
90 25 -300.00 0.00 -4.18
95 25 -300.00 0.00 2.03
100 25 -300.00 0.00 7.65
105 25 -300.00 0.00 12.62
110 25 -300.00 0.00 16.95
115 25 -300.00 0.00 20.67
120 25 -300.00 0.00 23.83
125 25 -300.00 0.00 26.51
130 25 -300.00 0.00 28.77
135 25 -300.00 0.00 30.65
140 25 -300.00 0.00 32.21
145 25 -300.00 0.00 33.48
150 25 -300.00 0.00 34.50
155 25 -300.00 0.00 35.30
160 25 -300.00 0.00 35.89
165 25 -300.00 0.00 36.30
170 25 -300.00 0.00 36.53
175 25 -300.00 0.00 36.60
180 25 -300.00 0.00 36.51
0 30 -300.00 0.00 -71.63
5 30 -300.00 0.00 -71.40
10 30 -300.00 0.00 -70.88
15 30 -300.00 0.00 -70.07
20 30 -300.00 0.00 -68.94
25 30 -300.00 0.00 -67.47
30 30 -300.00 0.00 -65.63
35 30 -300.00 0.00 -63.37
40 30 -300.00 0.00 -60.65
45 30 -300.00 0.00 -57.41
50 30 -300.00 0.00 -53.59
55 30 -300.00 0.00 -49.13
60 30 -300.00 0.00 -43.98
65 30 -300.00 0.00 -38.16
70 30 -300.00 0.00 -31.72
75 30 -300.00 0.00 -24.82
80 30 -300.00 0.00 -17.67
85 30 -300.00 0.00 -10.55
90 30 -300.00 0.00 -3.71
95 30 -300.00 0.00 2.62
100 30 -300.00 0.00 8.33
105 30 -300.00 0.00 13.35
110 30 -300.00 0.00 17.70
115 30 -300.00 0.00 21.43
120 30 -300.00 0.00 24.61
125 30 -300.00 0.00 27.28
130 30 -300.00 0.00 29.53
135 30 -300.00 0.00 31.40
140 30 -300.00 0.00 32.96
145 30 -300.00 0.00 34.22
150 30 -300.00 0.00 35.24
155 30 -300.00 0.00 36.03
160 30 -300.00 0.00 36.62
165 30 -300.00 0.00 37.03
170 30 -300.00 0.00 37.27
175 30 -300.00 0.00 37.34
180 30 -300.00 0.00 37.25
0 35 -300.00 0.00 -72.26
5 35 -300.00 0.00 -72.04
10 35 -300.00 0.00 -71.52
15 35 -300.00 0.00 -70.71
20 35 -300.00 0.00 -69.58
25 35 -300.00 0.00 -68.11
30 35 -300.00 0.00 -66.26
35 35 -300.00 0.00 -64.00
40 35 -300.00 0.00 -61.27
45 35 -300.00 0.00 -58.00
50 35 -300.00 0.00 -54.14
55 35 -300.00 0.00 -49.62
60 35 -300.00 0.00 -44.40
65 35 -300.00 0.00 -38.47
70 35 -300.00 0.00 -31.91
75 35 -300.00 0.00 -24.87
80 35 -300.00 0.00 -17.58
85 35 -300.00 0.00 -10.32
90 35 -300.00 0.00 -3.38
95 35 -300.00 0.00 3.04
100 35 -300.00 0.00 8.81
105 35 -300.00 0.00 13.87
110 35 -300.00 0.00 18.24
115 35 -300.00 0.00 21.98
120 35 -300.00 0.00 25.15
125 35 -300.00 0.00 27.83
130 35 -300.00 0.00 30.07
135 35 -300.00 0.00 31.94
140 35 -300.00 0.00 33.49
145 35 -300.00 0.00 34.76
150 35 -300.00 0.00 35.77
155 35 -300.00 0.00 36.57
160 35 -300.00 0.00 37.16
165 35 -300.00 0.00 37.57
170 35 -300.00 0.00 37.81
175 35 -300.00 0.00 37.89
180 35 -300.00 0.00 37.81
0 40 -300.00 0.00 -72.77
... 159721 more lines ...
|
| Attachment 4: XF_model_19_96.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -83.71 0.00 -158.06
5 0 -17.86 0.02 -153.75
10 0 -11.87 0.06 -148.66
15 0 -8.40 0.14 -144.50
20 0 -5.97 0.25 -140.71
25 0 -4.13 0.39 -136.31
30 0 -2.66 0.54 -131.91
35 0 -1.45 0.72 -127.38
40 0 -0.45 0.90 -123.42
45 0 0.39 1.09 -119.24
50 0 1.09 1.29 -115.23
55 0 1.69 1.47 -111.53
60 0 2.18 1.65 -107.68
65 0 2.59 1.81 -104.22
70 0 2.91 1.95 -100.95
75 0 3.15 2.07 -97.77
80 0 3.33 2.15 -94.90
85 0 3.43 2.20 -92.40
90 0 3.46 2.22 -89.68
95 0 3.43 2.20 -87.43
100 0 3.33 2.15 -85.51
105 0 3.15 2.07 -84.01
110 0 2.91 1.95 -82.20
115 0 2.59 1.81 -80.97
120 0 2.18 1.65 -79.70
125 0 1.69 1.47 -78.83
130 0 1.09 1.29 -77.85
135 0 0.39 1.09 -77.23
140 0 -0.45 0.90 -77.60
145 0 -1.45 0.72 -77.60
150 0 -2.66 0.54 -78.04
155 0 -4.13 0.39 -78.59
160 0 -5.97 0.25 -79.94
165 0 -8.40 0.14 -81.34
170 0 -11.87 0.07 -84.60
175 0 -17.86 0.02 -89.05
180 0 -83.70 0.00 -95.44
0 5 -83.74 0.00 -20.42
5 5 -17.86 0.02 -21.56
10 5 -11.87 0.06 -22.65
15 5 -8.40 0.14 -23.78
20 5 -5.97 0.25 -24.81
25 5 -4.13 0.39 -25.80
30 5 -2.66 0.54 -26.78
35 5 -1.45 0.72 -27.69
40 5 -0.45 0.90 -28.54
45 5 0.39 1.09 -29.38
50 5 1.09 1.29 -30.15
55 5 1.69 1.47 -30.88
60 5 2.18 1.65 -31.59
65 5 2.59 1.81 -32.25
70 5 2.91 1.95 -32.87
75 5 3.15 2.07 -33.45
80 5 3.33 2.15 -33.97
85 5 3.43 2.20 -34.45
90 5 3.46 2.22 -34.85
95 5 3.43 2.20 -35.18
100 5 3.33 2.15 -35.40
105 5 3.15 2.07 -35.52
110 5 2.91 1.95 -35.51
115 5 2.59 1.81 -35.38
120 5 2.18 1.65 -35.10
125 5 1.69 1.47 -34.70
130 5 1.09 1.29 -34.17
135 5 0.39 1.09 -33.48
140 5 -0.45 0.90 -32.71
145 5 -1.45 0.72 -31.84
150 5 -2.66 0.54 -30.83
155 5 -4.13 0.39 -29.77
160 5 -5.97 0.25 -28.61
165 5 -8.40 0.14 -27.42
170 5 -11.87 0.07 -26.17
175 5 -17.86 0.02 -24.90
180 5 -83.74 0.00 -23.60
0 10 -83.84 0.00 -19.91
5 10 -17.86 0.02 -20.98
10 10 -11.87 0.06 -22.04
15 10 -8.40 0.14 -23.08
20 10 -5.97 0.25 -24.09
25 10 -4.13 0.39 -25.07
30 10 -2.66 0.54 -26.00
35 10 -1.45 0.72 -26.89
40 10 -0.45 0.90 -27.75
45 10 0.39 1.09 -28.55
50 10 1.09 1.29 -29.32
55 10 1.69 1.47 -30.06
60 10 2.18 1.65 -30.76
65 10 2.59 1.81 -31.42
70 10 2.91 1.95 -32.05
75 10 3.15 2.07 -32.64
80 10 3.33 2.15 -33.17
85 10 3.43 2.20 -33.66
90 10 3.46 2.22 -34.07
95 10 3.43 2.20 -34.41
100 10 3.33 2.15 -34.64
105 10 3.15 2.07 -34.77
110 10 2.91 1.95 -34.79
115 10 2.59 1.81 -34.68
120 10 2.18 1.65 -34.44
125 10 1.69 1.47 -34.06
130 10 1.09 1.29 -33.55
135 10 0.39 1.09 -32.91
140 10 -0.45 0.90 -32.16
145 10 -1.45 0.72 -31.30
150 10 -2.66 0.54 -30.34
155 10 -4.13 0.39 -29.31
160 10 -5.97 0.25 -28.20
165 10 -8.40 0.14 -27.05
170 10 -11.87 0.07 -25.86
175 10 -17.86 0.02 -24.64
180 10 -83.84 0.00 -23.37
0 15 -84.01 0.00 -19.75
5 15 -17.86 0.02 -20.78
10 15 -11.87 0.06 -21.80
15 15 -8.40 0.14 -22.82
20 15 -5.97 0.25 -23.80
25 15 -4.13 0.39 -24.75
30 15 -2.66 0.54 -25.67
35 15 -1.45 0.72 -26.54
40 15 -0.45 0.90 -27.38
45 15 0.39 1.09 -28.17
50 15 1.09 1.29 -28.93
55 15 1.69 1.47 -29.66
60 15 2.18 1.65 -30.36
65 15 2.59 1.81 -31.01
70 15 2.91 1.95 -31.63
75 15 3.15 2.07 -32.21
80 15 3.33 2.15 -32.75
85 15 3.43 2.20 -33.22
90 15 3.46 2.22 -33.63
95 15 3.43 2.20 -33.96
100 15 3.33 2.15 -34.19
105 15 3.15 2.07 -34.33
110 15 2.91 1.95 -34.35
115 15 2.59 1.81 -34.24
120 15 2.18 1.65 -34.00
125 15 1.69 1.47 -33.65
130 15 1.09 1.29 -33.16
135 15 0.39 1.09 -32.54
140 15 -0.45 0.90 -31.82
145 15 -1.45 0.72 -30.99
150 15 -2.66 0.54 -30.07
155 15 -4.13 0.39 -29.06
160 15 -5.97 0.25 -28.00
165 15 -8.40 0.14 -26.87
170 15 -11.87 0.07 -25.71
175 15 -17.86 0.02 -24.52
180 15 -84.01 0.00 -23.30
0 20 -84.25 0.00 -19.66
5 20 -17.86 0.02 -20.66
10 20 -11.87 0.06 -21.65
15 20 -8.40 0.14 -22.63
20 20 -5.97 0.25 -23.59
25 20 -4.13 0.39 -24.52
30 20 -2.66 0.54 -25.42
35 20 -1.45 0.72 -26.27
40 20 -0.45 0.90 -27.10
45 20 0.39 1.09 -27.88
50 20 1.09 1.29 -28.63
55 20 1.69 1.47 -29.35
60 20 2.18 1.65 -30.03
65 20 2.59 1.81 -30.67
70 20 2.91 1.95 -31.28
75 20 3.15 2.07 -31.84
80 20 3.33 2.15 -32.36
85 20 3.43 2.20 -32.82
90 20 3.46 2.22 -33.21
95 20 3.43 2.20 -33.52
100 20 3.33 2.15 -33.75
105 20 3.15 2.07 -33.88
110 20 2.91 1.95 -33.89
115 20 2.59 1.81 -33.79
120 20 2.18 1.65 -33.57
125 20 1.69 1.47 -33.22
130 20 1.09 1.29 -32.76
135 20 0.39 1.09 -32.17
140 20 -0.45 0.90 -31.48
145 20 -1.45 0.72 -30.68
150 20 -2.66 0.54 -29.80
155 20 -4.13 0.39 -28.83
160 20 -5.97 0.25 -27.80
165 20 -8.40 0.14 -26.72
170 20 -11.87 0.07 -25.60
175 20 -17.86 0.02 -24.44
180 20 -84.25 0.00 -23.27
0 25 -84.56 0.00 -19.61
5 25 -17.86 0.02 -20.56
10 25 -11.87 0.06 -21.52
15 25 -8.40 0.14 -22.47
20 25 -5.97 0.25 -23.40
25 25 -4.13 0.39 -24.30
30 25 -2.66 0.54 -25.17
35 25 -1.45 0.72 -26.01
40 25 -0.45 0.90 -26.81
45 25 0.39 1.09 -27.58
50 25 1.09 1.29 -28.32
55 25 1.69 1.47 -29.01
60 25 2.18 1.65 -29.68
65 25 2.59 1.81 -30.31
70 25 2.91 1.95 -30.89
75 25 3.15 2.07 -31.44
80 25 3.33 2.15 -31.93
85 25 3.43 2.20 -32.37
90 25 3.46 2.22 -32.75
95 25 3.43 2.20 -33.04
100 25 3.33 2.15 -33.25
105 25 3.15 2.07 -33.37
110 25 2.91 1.95 -33.38
115 25 2.59 1.81 -33.28
120 25 2.18 1.65 -33.07
125 25 1.69 1.47 -32.74
130 25 1.09 1.29 -32.30
135 25 0.39 1.09 -31.75
140 25 -0.45 0.90 -31.09
145 25 -1.45 0.72 -30.35
150 25 -2.66 0.54 -29.50
155 25 -4.13 0.39 -28.58
160 25 -5.97 0.25 -27.60
165 25 -8.40 0.14 -26.56
170 25 -11.87 0.07 -25.49
175 25 -17.86 0.02 -24.38
180 25 -84.56 0.00 -23.25
0 30 -84.95 0.00 -19.57
5 30 -17.86 0.02 -20.48
10 30 -11.87 0.06 -21.40
15 30 -8.40 0.14 -22.31
20 30 -5.97 0.25 -23.20
25 30 -4.13 0.39 -24.07
30 30 -2.66 0.54 -24.92
35 30 -1.45 0.72 -25.73
40 30 -0.45 0.90 -26.51
45 30 0.39 1.09 -27.26
50 30 1.09 1.29 -27.97
55 30 1.69 1.47 -28.65
60 30 2.18 1.65 -29.30
65 30 2.59 1.81 -29.90
70 30 2.91 1.95 -30.47
75 30 3.15 2.07 -30.99
80 30 3.33 2.15 -31.45
85 30 3.43 2.20 -31.87
90 30 3.46 2.22 -32.21
95 30 3.43 2.20 -32.49
100 30 3.33 2.15 -32.69
105 30 3.15 2.07 -32.79
110 30 2.91 1.95 -32.80
115 30 2.59 1.81 -32.71
120 30 2.18 1.65 -32.51
125 30 1.69 1.47 -32.20
130 30 1.09 1.29 -31.79
135 30 0.39 1.09 -31.27
140 30 -0.45 0.90 -30.66
145 30 -1.45 0.72 -29.95
150 30 -2.66 0.54 -29.16
155 30 -4.13 0.39 -28.30
160 30 -5.97 0.25 -27.37
165 30 -8.40 0.14 -26.39
170 30 -11.87 0.07 -25.36
175 30 -17.86 0.02 -24.31
180 30 -84.96 0.00 -23.23
0 35 -85.43 0.00 -19.55
5 35 -17.86 0.02 -20.41
10 35 -11.87 0.06 -21.27
15 35 -8.40 0.14 -22.14
20 35 -5.97 0.25 -22.99
25 35 -4.13 0.39 -23.82
30 35 -2.66 0.54 -24.63
35 35 -1.45 0.72 -25.42
40 35 -0.45 0.90 -26.17
45 35 0.39 1.09 -26.90
50 35 1.09 1.29 -27.58
55 35 1.69 1.47 -28.24
60 35 2.18 1.65 -28.86
65 35 2.59 1.81 -29.44
70 35 2.91 1.95 -29.98
75 35 3.15 2.07 -30.47
80 35 3.33 2.15 -30.91
85 35 3.43 2.20 -31.30
90 35 3.46 2.22 -31.61
95 35 3.43 2.20 -31.87
100 35 3.33 2.15 -32.05
105 35 3.15 2.07 -32.14
110 35 2.91 1.95 -32.15
115 35 2.59 1.81 -32.06
120 35 2.18 1.65 -31.88
125 35 1.69 1.47 -31.60
130 35 1.09 1.29 -31.22
135 35 0.39 1.09 -30.74
140 35 -0.45 0.90 -30.17
145 35 -1.45 0.72 -29.52
150 35 -2.66 0.54 -28.79
155 35 -4.13 0.39 -27.98
160 35 -5.97 0.25 -27.11
165 35 -8.40 0.14 -26.19
170 35 -11.87 0.07 -25.23
175 35 -17.86 0.02 -24.23
180 35 -85.44 0.00 -23.22
0 40 -86.01 0.00 -19.53
... 159721 more lines ...
|
| Attachment 5: XF_model_29_87.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -77.59 0.00 -23.13
5 0 -17.30 0.02 -23.13
10 0 -11.30 0.07 -23.11
15 0 -7.83 0.16 -23.07
20 0 -5.40 0.29 -23.00
25 0 -3.56 0.44 -22.93
30 0 -2.09 0.62 -22.84
35 0 -0.89 0.82 -22.73
40 0 0.11 1.03 -22.61
45 0 0.95 1.24 -22.47
50 0 1.66 1.46 -22.33
55 0 2.25 1.68 -22.17
60 0 2.74 1.88 -22.01
65 0 3.14 2.06 -21.84
70 0 3.46 2.22 -21.66
75 0 3.71 2.35 -21.48
80 0 3.88 2.44 -21.29
85 0 3.99 2.50 -21.11
90 0 4.02 2.52 -20.92
95 0 3.99 2.50 -20.74
100 0 3.88 2.45 -20.55
105 0 3.71 2.35 -20.38
110 0 3.47 2.22 -20.20
115 0 3.15 2.06 -20.03
120 0 2.74 1.88 -19.87
125 0 2.25 1.68 -19.72
130 0 1.66 1.47 -19.58
135 0 0.96 1.25 -19.44
140 0 0.12 1.03 -19.31
145 0 -0.88 0.82 -19.20
150 0 -2.08 0.62 -19.10
155 0 -3.55 0.44 -19.02
160 0 -5.39 0.29 -18.94
165 0 -7.82 0.17 -18.89
170 0 -11.29 0.07 -18.84
175 0 -17.28 0.02 -18.82
180 0 -77.55 0.00 -18.81
0 5 -77.73 0.00 -18.36
5 5 -17.30 0.02 -18.44
10 5 -11.30 0.07 -18.54
15 5 -7.83 0.16 -18.64
20 5 -5.40 0.29 -18.77
25 5 -3.56 0.44 -18.92
30 5 -2.09 0.62 -19.09
35 5 -0.89 0.82 -19.27
40 5 0.11 1.03 -19.47
45 5 0.95 1.24 -19.67
50 5 1.66 1.46 -19.89
55 5 2.25 1.68 -20.12
60 5 2.74 1.88 -20.35
65 5 3.14 2.06 -20.60
70 5 3.46 2.22 -20.84
75 5 3.71 2.35 -21.08
80 5 3.88 2.44 -21.33
85 5 3.99 2.50 -21.57
90 5 4.02 2.52 -21.81
95 5 3.99 2.50 -22.04
100 5 3.88 2.45 -22.28
105 5 3.71 2.35 -22.50
110 5 3.47 2.22 -22.72
115 5 3.15 2.06 -22.92
120 5 2.74 1.88 -23.12
125 5 2.25 1.68 -23.30
130 5 1.66 1.47 -23.47
135 5 0.96 1.25 -23.62
140 5 0.12 1.03 -23.75
145 5 -0.88 0.82 -23.85
150 5 -2.08 0.62 -23.94
155 5 -3.55 0.44 -24.00
160 5 -5.39 0.29 -24.04
165 5 -7.82 0.17 -24.06
170 5 -11.29 0.07 -24.04
175 5 -17.28 0.02 -24.01
180 5 -77.69 0.00 -23.95
0 10 -77.94 0.00 -16.18
5 10 -17.30 0.02 -16.28
10 10 -11.30 0.07 -16.44
15 10 -7.83 0.16 -16.61
20 10 -5.40 0.29 -16.84
25 10 -3.56 0.44 -17.09
30 10 -2.09 0.62 -17.37
35 10 -0.89 0.82 -17.69
40 10 0.11 1.03 -18.03
45 10 0.95 1.24 -18.40
50 10 1.66 1.46 -18.78
55 10 2.25 1.68 -19.19
60 10 2.74 1.88 -19.60
65 10 3.14 2.06 -20.03
70 10 3.46 2.22 -20.46
75 10 3.71 2.35 -20.89
80 10 3.88 2.44 -21.33
85 10 3.99 2.50 -21.76
90 10 4.02 2.52 -22.19
95 10 3.99 2.50 -22.61
100 10 3.88 2.45 -23.02
105 10 3.71 2.35 -23.43
110 10 3.47 2.22 -23.82
115 10 3.15 2.06 -24.20
120 10 2.74 1.88 -24.55
125 10 2.25 1.68 -24.88
130 10 1.66 1.47 -25.19
135 10 0.96 1.25 -25.46
140 10 0.12 1.03 -25.71
145 10 -0.88 0.82 -25.92
150 10 -2.08 0.62 -26.09
155 10 -3.55 0.44 -26.22
160 10 -5.39 0.29 -26.30
165 10 -7.82 0.17 -26.35
170 10 -11.29 0.07 -26.36
175 10 -17.28 0.02 -26.33
180 10 -77.89 0.00 -26.25
0 15 -78.23 0.00 -14.91
5 15 -17.30 0.02 -15.04
10 15 -11.30 0.07 -15.22
15 15 -7.83 0.16 -15.44
20 15 -5.40 0.29 -15.71
25 15 -3.56 0.44 -16.02
30 15 -2.09 0.62 -16.37
35 15 -0.89 0.82 -16.76
40 15 0.11 1.03 -17.19
45 15 0.95 1.24 -17.64
50 15 1.66 1.46 -18.12
55 15 2.25 1.68 -18.63
60 15 2.74 1.88 -19.14
65 15 3.14 2.06 -19.68
70 15 3.46 2.22 -20.21
75 15 3.71 2.35 -20.76
80 15 3.88 2.44 -21.30
85 15 3.99 2.50 -21.85
90 15 4.02 2.52 -22.39
95 15 3.99 2.50 -22.92
100 15 3.88 2.45 -23.44
105 15 3.71 2.35 -23.94
110 15 3.47 2.22 -24.43
115 15 3.15 2.06 -24.91
120 15 2.74 1.88 -25.35
125 15 2.25 1.68 -25.77
130 15 1.66 1.47 -26.16
135 15 0.96 1.25 -26.51
140 15 0.12 1.03 -26.82
145 15 -0.88 0.82 -27.09
150 15 -2.08 0.62 -27.31
155 15 -3.55 0.44 -27.48
160 15 -5.39 0.29 -27.60
165 15 -7.82 0.17 -27.67
170 15 -11.29 0.07 -27.68
175 15 -17.28 0.02 -27.65
180 15 -78.17 0.00 -27.56
0 20 -78.59 0.00 -14.07
5 20 -17.30 0.02 -14.21
10 20 -11.30 0.07 -14.41
15 20 -7.83 0.16 -14.66
20 20 -5.40 0.29 -14.95
25 20 -3.56 0.44 -15.30
30 20 -2.09 0.62 -15.70
35 20 -0.89 0.82 -16.14
40 20 0.11 1.03 -16.61
45 20 0.95 1.24 -17.12
50 20 1.66 1.46 -17.66
55 20 2.25 1.68 -18.23
60 20 2.74 1.88 -18.82
65 20 3.14 2.06 -19.42
70 20 3.46 2.22 -20.03
75 20 3.71 2.35 -20.64
80 20 3.88 2.44 -21.26
85 20 3.99 2.50 -21.88
90 20 4.02 2.52 -22.49
95 20 3.99 2.50 -23.09
100 20 3.88 2.45 -23.69
105 20 3.71 2.35 -24.26
110 20 3.47 2.22 -24.82
115 20 3.15 2.06 -25.36
120 20 2.74 1.88 -25.87
125 20 2.25 1.68 -26.34
130 20 1.66 1.47 -26.78
135 20 0.96 1.25 -27.18
140 20 0.12 1.03 -27.53
145 20 -0.88 0.82 -27.84
150 20 -2.08 0.62 -28.09
155 20 -3.55 0.44 -28.29
160 20 -5.40 0.29 -28.43
165 20 -7.82 0.17 -28.52
170 20 -11.29 0.07 -28.54
175 20 -17.28 0.02 -28.51
180 20 -78.53 0.00 -28.42
0 25 -79.03 0.00 -13.47
5 25 -17.30 0.02 -13.62
10 25 -11.30 0.07 -13.83
15 25 -7.83 0.16 -14.09
20 25 -5.40 0.29 -14.41
25 25 -3.56 0.44 -14.78
30 25 -2.09 0.62 -15.20
35 25 -0.89 0.82 -15.67
40 25 0.11 1.03 -16.18
45 25 0.95 1.24 -16.73
50 25 1.66 1.46 -17.31
55 25 2.25 1.68 -17.92
60 25 2.74 1.88 -18.55
65 25 3.14 2.06 -19.20
70 25 3.46 2.22 -19.86
75 25 3.71 2.35 -20.53
80 25 3.88 2.44 -21.20
85 25 3.99 2.50 -21.87
90 25 4.02 2.52 -22.54
95 25 3.99 2.50 -23.20
100 25 3.88 2.45 -23.84
105 25 3.71 2.35 -24.47
110 25 3.47 2.22 -25.08
115 25 3.15 2.06 -25.66
120 25 2.74 1.88 -26.21
125 25 2.25 1.68 -26.73
130 25 1.66 1.47 -27.21
135 25 0.96 1.25 -27.64
140 25 0.12 1.03 -28.03
145 25 -0.88 0.82 -28.36
150 25 -2.08 0.62 -28.64
155 25 -3.55 0.44 -28.86
160 25 -5.40 0.29 -29.02
165 25 -7.82 0.17 -29.12
170 25 -11.29 0.07 -29.15
175 25 -17.28 0.02 -29.13
180 25 -78.97 0.00 -29.04
0 30 -79.57 0.00 -13.01
5 30 -17.30 0.02 -13.16
10 30 -11.30 0.07 -13.37
15 30 -7.83 0.16 -13.65
20 30 -5.40 0.29 -13.97
25 30 -3.56 0.44 -14.36
30 30 -2.09 0.62 -14.80
35 30 -0.89 0.82 -15.29
40 30 0.11 1.03 -15.82
45 30 0.95 1.24 -16.40
50 30 1.66 1.46 -17.01
55 30 2.25 1.68 -17.65
60 30 2.74 1.88 -18.32
65 30 3.14 2.06 -19.00
70 30 3.46 2.22 -19.70
75 30 3.71 2.35 -20.41
80 30 3.88 2.44 -21.12
85 30 3.99 2.50 -21.84
90 30 4.02 2.52 -22.55
95 30 3.99 2.50 -23.24
100 30 3.88 2.45 -23.93
105 30 3.71 2.35 -24.60
110 30 3.47 2.22 -25.25
115 30 3.15 2.06 -25.87
120 30 2.74 1.88 -26.46
125 30 2.25 1.68 -27.01
130 30 1.66 1.47 -27.52
135 30 0.96 1.25 -27.98
140 30 0.12 1.03 -28.39
145 30 -0.88 0.82 -28.75
150 30 -2.08 0.62 -29.05
155 30 -3.55 0.44 -29.28
160 30 -5.40 0.29 -29.46
165 30 -7.82 0.17 -29.57
170 30 -11.29 0.07 -29.61
175 30 -17.28 0.02 -29.59
180 30 -79.50 0.00 -29.50
0 35 -80.22 0.00 -12.65
5 35 -17.30 0.02 -12.80
10 35 -11.30 0.07 -13.01
15 35 -7.83 0.16 -13.28
20 35 -5.40 0.29 -13.62
25 35 -3.56 0.44 -14.02
30 35 -2.09 0.62 -14.47
35 35 -0.89 0.82 -14.97
40 35 0.11 1.03 -15.52
45 35 0.95 1.24 -16.11
50 35 1.66 1.46 -16.75
55 35 2.25 1.68 -17.41
60 35 2.74 1.88 -18.10
65 35 3.14 2.06 -18.81
70 35 3.46 2.22 -19.54
75 35 3.71 2.35 -20.28
80 35 3.88 2.44 -21.03
85 35 3.99 2.50 -21.78
90 35 4.02 2.52 -22.52
95 35 3.99 2.50 -23.26
100 35 3.88 2.45 -23.98
105 35 3.71 2.35 -24.68
110 35 3.47 2.22 -25.36
115 35 3.15 2.06 -26.02
120 35 2.74 1.88 -26.63
125 35 2.25 1.68 -27.21
130 35 1.66 1.47 -27.75
135 35 0.96 1.25 -28.23
140 35 0.12 1.03 -28.66
145 35 -0.88 0.82 -29.04
150 35 -2.08 0.62 -29.36
155 35 -3.55 0.44 -29.61
160 35 -5.40 0.29 -29.80
165 35 -7.82 0.17 -29.92
170 35 -11.29 0.07 -29.97
175 35 -17.28 0.02 -29.96
180 35 -80.14 0.00 -29.87
0 40 -80.99 0.00 -12.34
... 159721 more lines ...
|
|
81
|
Thu Jul 2 23:13:05 2020 |
Amy | A useful reference on bicone antennas |
I found this datasheet on bicone antennas, that is a useful reference for seeing the relationship between bicone dimensions and bandwidth. From these it looks like the lower end of the band is at about f_low=c/(4*L_E), where L_E is the entire length of the antenna (twice the length of one cone), although we'd expect the peak gain to be where a quarter wavelength is just one of the cones (and the figure may roughly support that).
The attached file comes from this website:
https://www.ramayes.com/Biconical_Antennas.htm
|
| Attachment 1: biconall.pdf
|
|
180
|
Thu Aug 18 10:32:17 2022 |
Amy | reading about HPol antennas |
Here are some reading materials about HPol antennas:
This paper discusses some initial HPol designs that were tested in a prototype station of ARA. We went with the slotted cylinder design.
Here you can see what the RNO-G experiment in Greenland has considered.
We can add other sources here as we find them. |
|
202
|
Mon Mar 20 12:37:38 2023 |
Amy | ANITA/PUEO pictures |
Here are some ANITA/PUEO pictures that I received from Christian Mike that might be helpful. I include his descriptions as well.
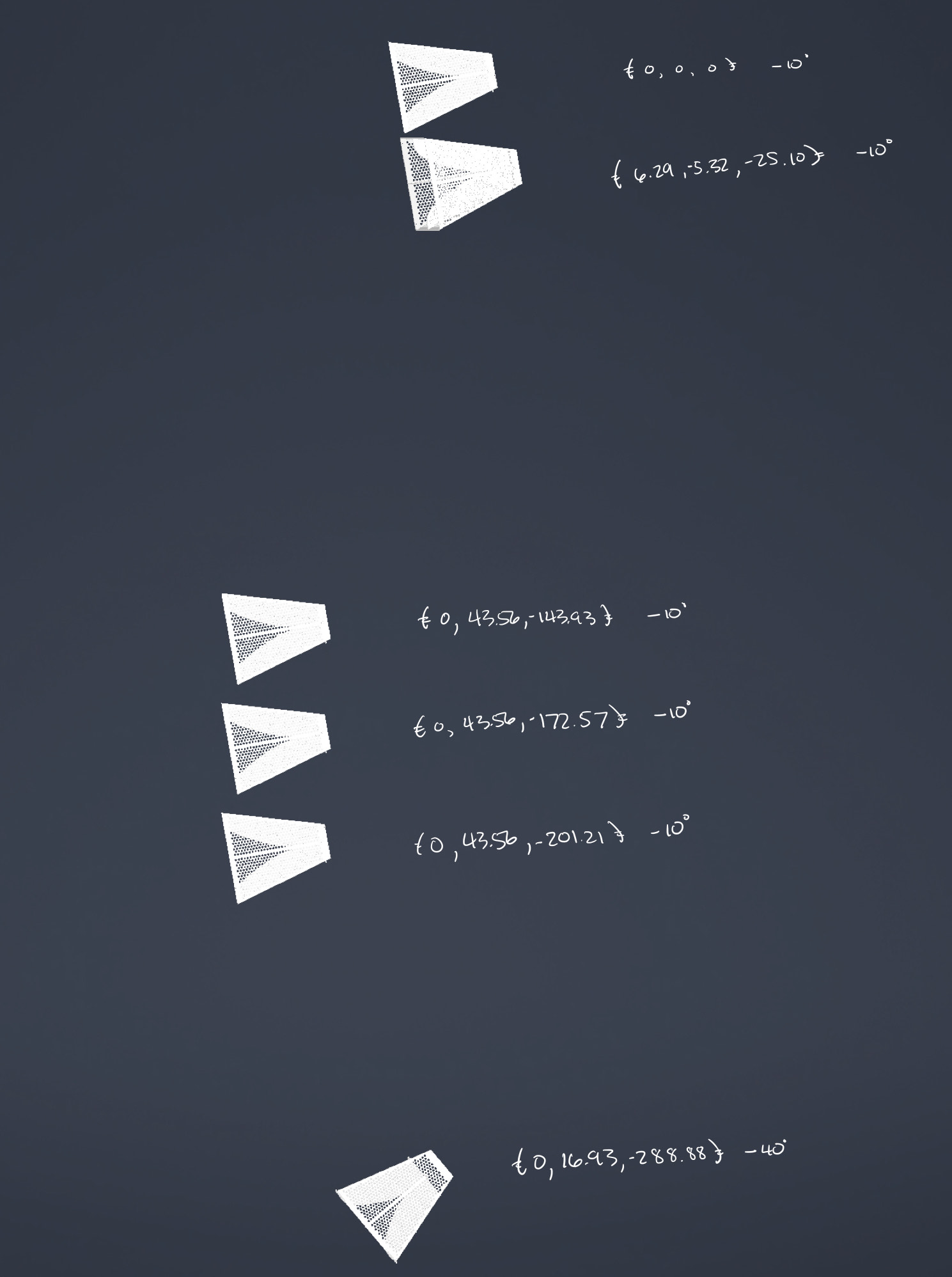
The one with the black crates in the background is the ANITA 1 and ANITA 2 form factor and the one in the anechoic chamber is the ANITA 3 and ANITA 4 form factor. I dimensioned out an ANITA 3/4 antenna, in CAD space, to give you a better idea of the ridge profile. The ANITA 1/2 CAD models we have, I found, are inaccurate.
Attached are images of the PUEO antenna array geometry. One image annotates the approximate locations of the Vpol feed with the top antenna feed as the origin.
|
| Attachment 1: IMG_2362.jpg
|
 |
| Attachment 2: IMG_2364.jpg
|
 |
| Attachment 3: IMG_2365.jpg
|
 |
|
203
|
Mon Mar 20 13:35:25 2023 |
Amy | info on format for PUEO antenna files |
William Luszczak 1:30 PM
This is the directory with the current PUEO antenna data files: https://github.com/PUEOCollaboration/pueoSim/tree/main/data/antennas. Each type of antenna will need several files:
- vv_0 and hh_0 describe the on axis v and h pol gain as a function of frequency. Gain is listed as a multiplicative factor (not in dB, if I'm remembering correctly)
- hv_0 and vh_0 describe the on axis cross polarization gain
- vv_az and hh_az describe the off-axis gain via multiplicative factors of the boresight gain. These are currently listed as a function of frequency for reference angles of 0,10,20,30,40,50,60,70,80, and 90 degrees, though we can always adjust the reference angles if needed.
- vv_el and hh_el are similar to above, but for gain as a function of elevation angle instead of azimuthal angle.
So for example, the total vpol gain in a particular frequency bin for a signal incident at azimuthal angle az and elevation angle el would be vv_0(f)*vv_az(f, az)*vv_el(f, el)
New
1:31
We can also potentially adjust this if it would be more convenient for the GENETIS people to output antenna information in a different format (or at different reference angles or whatever). The important thing is that the boresight h, v, and cross pol gains are all defined, as well as the off-axis response (as a function of azimuth and elevation) |
|
219
|
Mon May 22 20:34:59 2023 |
Amy | Things to do before starting new PUEO run from 5/22/23 meeting |
Here is a list of things we agreed to do before starting a new PUEO run. Feel free to add notes here as things are done and we will revisit the list next week.
Change range of the height variable to 0.75-1.75m
Start with all bad individuals so that we can be sure and see growth - all at 0.75m?
Make sure we are using the ratios in the GA that we think we are
If we are using reproduction, are we combining results when individuals are resimulated to reduce errors? I made a program to accomplish this and created a pull request on the PUEO Github (Dylan)
Investigate whether there are obvious places to speed up PUEOSim
Consolidate the GA to one branch
Understand what is happening with cross-pol, make sure we're not gettting Veff=0 when using XF-generated cross-pol response
Edit: Have the xmacros use the realized gain instead of the idealized gain, which allows for the design to build in matching
|
|
237
|
Wed Aug 2 22:33:18 2023 |
Amy | slides from building meeting 8/2/23 |
Slides shown at experts building meeting Aug. 2nd, 2023. |
| Attachment 1: GENETIS_building_080223.pdf
|
|
238
|
Mon Aug 21 15:47:13 2023 |
Amy | OSC license agreement to be able to use XF |
Attached is the license agreement that each person should sign to be able to use XF on OSC. You can sign it, send it to Amy, and she will return it to OSC with her signature on it. |
| Attachment 1: User_Software_Agreement-1.pdf
|
| Attachment 2: xfdtd.pdf
|
|
14
|
Mon Jul 1 13:45:49 2019 |
Alex Patton | Status on optimizing AraSim |
We ran valgrind (a profiler) on AraSim to find out where it was spending most of its time, we ran it with callgrind to give an output file of the time and opened that file with qcashegrind to display it in a legible format. This let us know that the function Param_RE_Tterm_approx was taking up about 75% of the time because it was being called hundreds of millions of times. Now more specifically the standard math functions pow() and exp() were being called in this function every time and took up about 44% and 20% respectively. They were called about the same number of times as Param_RE_Tterm_approx but they are only called in the else block in an if-else statement.
JULY 31st Update:
I used the C++ library future and set up iterations of a loop to run asynchronously as they do not rely on each other's calculations. This launches a seperate thread whenever I call one. The struggle with this is that launching a thread and getting its results takes time as well, so there had to be enough calulations in a thread in order for it to make up any time. The purpose of the function GetVm_FarField_Tarray is to create arrays of doubles called Earray and Tarray and those are the only values that get altered in this function. Tarray is created with simple calculations so I left that out of threads but making Earray calls the other function that takes up so much time.
I edited the files signal.cc and signal.hh to work with 4 threads and ran 10,000 neutrinos with 4 cores. The cpu time was 1:48:41 but the real walltime it used only took 00:54:40. The time these tests for 10,000 neutrinos with base AraSim normally take anywhere from 1:35:00 to 1:45:00 or in that range. It should still work with less threads available it would just run slower and probably at the cpu time, which might be slightly slower than base AraSim but as soon as more threads are available it runs much faster as we see here. I still need to check for the accuracy of these edits, the number passed is in a reasonable range, but I will need to investigate further by giving the exact same neutrinos to both programs and making sure their outputs are identical.
I also have some further ideas to speed it up even more. Currently every iteration of the loop calls a thread but what would be faster would be to either
A: Set it up so it can call more threads at a time and thus go faster, which would be an easy edit but only work if more cores are given
B: Set it up so the current threads run more than one iteration at a time, I've started experimenting with this but having trouble making sure that everything involving pointers stays safe, but the benefit of this is that the threads would get called and joined significantly less and would cause a great speed increase without adding more threads.
All these developments are new after me spending a while trying many different ways to speed it up or run parts in parallel like this, so even if this isn't accurate or valid I believe I would be able to make it so with more time and work.
Attached I have a zip that contains the two files I edited as well as an output file from my first test. I will continue to test and improve this but am unable to work on this again until August 19th so I wanted to share my current status before I am unavailable. |
| Attachment 1: NewFilesAndResults.zip
|
|
50
|
Tue May 19 12:20:40 2020 |
Alex Patton | GENETIS Daily Updates |
Today's Summer 2020 daily update:
As a note, today OSC was down so productivity was more limited
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Mostly just wrote more on the paper in the Genetic Algorithm section. I added some citation that we used in ICRC but there are still more places that should have citations.
|
I might check tonight when OSC is back up to try to push in more updates to the loop because I wanna get Evelyn and Ryan started on running the loop. Putting in those fixes is a big priority because we want to be able to correct the potential issue with XF simulation folders being overwritten and thus uan data not corresponding to the write individuals. The two big things for me in the loop are getting the simulation data to save correctly (and also putting that in the database) and testing that we can replicate results using the specific seed. I'll probably only focus on loop stuff tomorrow.
|
| Alex P. |
Got up before OSC was down to check progress of overnight run, it seems to have worked but I noticed a problem with the database that it wasn't writing to it probably due to a permissions issue but I would have to run another time to see. Shouldn't have affected data but just the use of the database. Run got up to 8 generations with non-zero fitness score which is positive and seems to have fixed the error we originally encountered. Talked to Eliot about pointers and possible errors but was unable to look at the specific error because it is on OSC.
|
Tomorrow plan is to continue to work on database functionality and continue run to get more generations, also want to add the ability to add more plots than just the fitness score to the dropbox automatically. Plots: upload all plots (Fitness, LRT, vEff), remove legend, upload penalized red/green plot too, take off legend, add units to Fitness |
| Leo |
|
|
| Eliot |
Read about pointers and vectors in C++. Talked to Alex P a bit, and have some ideas of things to change to get the GA running. Began reading about antennas. Mostly a down day due to OSC being down.
|
Will implement changes to GA and continue familiarizing myself with how XF reads these values.
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
52
|
Wed May 20 10:59:29 2020 |
Alex Patton | GENETIS Daily Update |
Today's Summer 2020 daily update:
Learn how to use the final part of the loop
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Currently running the loop without the database to change the fix Alex and I made for XF to properly place data in the simulation folders. It looks to us like this was a problem that needed to be fixed. I'm running with the specific seed set in the roulette algorithm so we can replicate the results. I showed Evelyn and Ryan how to run the code today. Ryan was able to get a Pitzer Desktop, so until we can figure out how to get his X11 forwarding to work for XF, that will be a temporary fix. The only drawback is that Pitzer desktops require a GPU, so wait times can be high. I started this run by having him screenshare on zoom and then guiding him through running the loop. We got through the submission of the first batch of XF jobs (and also found some errors, so we had a chance to demonstrate fixing those).
|
My goal is to get several generations of this run done so that I have enough data to see if my fix worked. If so, I'm going to implement it in the database version and we'll perform a test with that. I'm meeting with Evelyn and Ryan at 1:30 to discuss the loop again, so I'll try to have a run going near where we left off today so they can see the rest. Once we know that we've fixed this XF problem we should be ready to fully implement the database by merging with the master branch.
|
| Alex P. |
Implemented some changes to get graphs working and automatically submitted all 5 graphs requested for to dropbox, then helped Alex M debug his changes and helped him show Evelyn and Ryan the loop. While Alex M ran to test the changes, helped Eliot and Leo with their pointer problem and fixed it so it doesn't give an error but they still need to make some changes to get it functioning how they want. |
Continue to help Alex M with fixing the XF simulationNumber issue and make sure the graphs are all valid and uploading properly. After we get confirmation that our XF fix is working we will implement it to the database version too. |
| Leo |
Me and Eliot continued work on the 2 chromosome method. We were successful at fixing our pointing/vector issue, but we have a new error with the data_read function within the GA. |
Tomorrow we will meet with Julie to flush out the issue with data_read. |
| Eliot |
Continued working on the erros with the roulette alg. I have gotten the code to a running state, however the data is not outputting correctly. I believe this is an issue within the roulette function or dataread function.
|
I will further investigate roulette and dataread function and meet with Julie to do so.
|
| Evelyn |
Learned how to run the updated version of the loop and about the database, but we weren't able to get through an entire generation.
|
Learn how to use the final part of the loop. |
| Ryan |
I found a workaround (using a Pitzer desktop until I can fix the error with my machine's Linux subsystem) to my previous issue and was able to do two runs of the loop. |
Alex M. Plans on helping me finish learning how to run the loops.
|
|
|
53
|
Thu May 21 14:31:08 2020 |
Alex Patton | GENETIS Daily Update |
Today's Summer 2020 daily update:
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Continued the run testing the XF fixes with Alex P. We think our fixes are working, but we want to get a few more generations through. We checked the uan files and saw different data, which we didn't see on the past runs, so this indicates the fix is working. Alex and I also showed Ryan and Evelyn more of the loop and how to interpret the plots being generated. I also put in the red-green color plots, but we need to get through the next generation to see if they work.
|
I'm going to try to get through more generations tonight so we can see what other fixes we still need to implement. I'll continue looking at any bugs that arise tomorrow, but we're getting very close to being able to mergre branches.
|
| Alex P. |
Picked up Ryan and Alex's run from yesterday to progress to the end of gen 1. Found an error related to the changes and moving UAN files after gen 0 with proper naming. After error was fixed went back to run and made sure the graphs all worked. XFintoARA.py had to be altered to fit the new changes. We saw that the UAN files now differed between individuals across generations whereas they hadn't before. Worked with Alex M and showed Evelyn and Ryan more about the loop and the graphs while also debugging our problems while sharing with them so they can see the process when running the loop and running into an error. |
Continue to run, hoping to iron out all errors so it can run on its own. Along with that making sure all the graph tools work, currently have an error in the Red/Green penalized version and also in the automatic upload of some graphs. Also hope to continue the old database run past generation 9 before implementing these changes on the database version. |
| Leo |
|
|
| Eliot |
Font Continued to work on erros within roulette algorithm. Met with Julie to discuss the issue we were still having. Soon after meeting, Leo and I got it to a near perfect state. There are only a couple genese that were unique that shouldn't have been. This is likely a small error that I will look into tomorrow.
|
I plan to finalize the roulette alg to accept 2 chromosomes with 3 genes a piece. These will have shared radius and theta but unique lengths.
|
| Evelyn |
Learned how to run, debug, and read the graphs in the second half of the loop, so that I can start getting some real data as soon as the loop is ready to be run again. |
|
| Ryan |
|
|
|
|
56
|
Wed May 27 15:00:28 2020 |
Alex Patton | Daily ELOG Update 5/27/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Met with Alex P. and Amy to think about how we can make sure the data coming from AraSim makes sense (specifically, to figure out why we're getting effective volumes of 0 on some antennas). Amy suggested we use a function in AraSim that let's us simulate identical events so that we can compare the same events between two different antennas. I asked Jorge in the #general chat to see if he could point us to where to look in AraSim for that function.
Met with Amy and Julie about people interested in joining the group and what upcoming project we have. We also talked about Karoo and Kai's thesis and how we can use machine learning to classify noise in the Anita data.
|
I won't be able to meet tomorrow or Friday morning, but I'll try to jump on in the afternoon on Friday and I should be able to make the GENETIS call. Also, Julie wanted to find a time where we could both meet with one of the new members if they have time this week. |
|
Alex P.
|
Worked in morning with Alex trying to find AraSim's 0 vEff problem, set it up to save AraOut files from now on in Run_Outputs in order to more easily look through. Continued with Git and successfully have working copies set up across multiple people and branches so we can all work without any conflicts. |
Continue to run and try to debug. Next idea is probably to set it up to pass exact events as specified so we know exactly what event is running and find error that way. |
| Eliot |
Today we finalized the GA. It now works with 2 chromosomes, where only length varies across chromosomes. In addition we got the tournament selection working (we don’t think it was working for even 1 chromosome prior). It is ready to begin being implemented tomorrow. We also met with Julie and others to discuss git and related topics.
|
Tomorrow we will implement the GA and begin editing Xmacros to 1) tell XF the new antenna type, etc and 2) have it read the new 2 chromosome data properly. This will likely take multiple days.
|
| Leo |
|
|
| Evelyn |
Learned more about git and github from Julie |
Find the old paperclip code and learn how to run and how it works! |
| Ryan |
Continued learning about Git and Github uses from Julie. Went back in the elog history to learn more about a previous "paperclip algorithm" for future uses. |
Learn more about the paperclip algorithm ad possibly help with more runs. |
|
|
Draft
|
Thu May 28 15:11:35 2020 |
Alex Patton | GENETIS Daily Update 5/28/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
I wasn't available Thursday or Friday morning.
|
My goal on Friday is to help Alex P. more with finding the cause of the 0 effective volumes we're getting (and also the high effective volumes for small antennas). |
|
Alex P.
|
Set up AraSim to use event read in, got 100 events generated from AraSim in the format of an EventReadIn file for AraSim so that way we have events that are realistic. Tried running and got a run up to the AraSim phase but had some problems with the proper call and order of argumetns. I looked through the AraOut files and thought I got it set up but got some "cannot open" errors I will deal with later. Then hopped on call with Julie, Eliot, and Leo to deal with a Git conflict |
Follow up on AraSim read in to get it functioning properly and then make sure to find events that pass for good antennas so that we can find errors in our 0 vEff antennas. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
d |
|
59
|
Mon Jun 1 15:21:03 2020 |
Alex Patton | Daily ELOG Update 6/1/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Spoke with Evelyn and Ryan about the paperclip antennas. They found the github repository with the files and a README so they're getting started on learning about the algorithm and testing out the scripts. Once they play with it and can get it running they'll start running it with different parameters for the selection methods (roulette vs tournament).
Also worked with Alex P. on figuring out the issue with the effective volumes coming out of AraSim. We have data from the eventReadIn function in AraSim for the small antennas and we don't see differences between the gains in these antennas and the actual bicone, but we still need to look at more of the quantities to compare that Amy told us about last week. We also came up with a new plot that might be interesting to look at. We think the plots look suspicious--I think it will be worthwhile to try rerunning with the same antennas but using a smaller grid spacing to see if the shape of the plots drastically changes.
|
Going to keep working with Alex P on resolving the small antenna effective volume issue. I'm also going to meet with Julie and Ben at 1 since Julie couldn't meet today. |
|
Alex P.
|
Continued to test with the Ara 0 vEff problem. Ran individuals with feed through events and individuals that gave 0 vEff had zero individuals pass still where others had consistent number pass. Used diff command and looked at AraOutput files and saw no difference except saying which events passed. Difference lies in .dat files passed into them which are generated from uan. These files are 130,000+ lines so we made a program to graph that average gain over the frequencies for the individuals and ran it with our small antennas. Interestingly, the ones that failed had higher average gains consistently than some runs that passed. Also ran this program with the Actual Bicone and the results were pretty different from the bicone and these small antenna. |
Next plan is to run our graphing code on a generation of previously large antennas generated to see if it makes any difference and maybe the problem is how these small antennas fitness scores get generated. |
| Eliot & Leo |
|
|
| Evelyn |
We were able to dig up the old paperclip code and make a new github repository with those files so we can work on it without breaking the originals. |
Figure out how to run the paperclip code from our own machines |
| Ryan |
Evelyn and I found the old paperclip repository on git, copied it over to our machines, and created a new repository to do our work on for the future. |
Create a dev branch and figure out how to run the algorithm. |
|
|
60
|
Tue Jun 2 13:42:21 2020 |
Alex Patton | Daily GENETIS Update 6/2/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Worked with Alex on trying to compare bicones to the ARA bicone. Met with Ben to give him some things to look over and told him to come to the meeting tomorrow at 1.
|
I might not be available all day tomorrow, but I should be good to come to the meeting at 1. Planning on doing a little bit of review of neural networks to get a feel of some of the stuff Julie is doing with Kai--I'm looking over notes and assignments from my ML class last fall. I also have a few things in the paper to fix up (namely citations, which I haven't been able to get to work). |
|
Alex P.
|
Ran analysis with bigger antennas and saw that they match the actual bicone more than the smaller ones in their overall average gain patterns, reduced grid spacing and started a run to see if it will change the outputs for the small antennas since we know that XF's generation of the UAN is the difference between gains and individuals. |
Observe results from the reduced grid spacing run and hopefully find some enlighting information to determine the next step. If not will try to find how to output electric field and compare that between our trials |
| Eliot & Leo |
|
|
| Evelyn |
We were able to find and run the old paperclip genetic algorithms, but we aren't completely sure how they work or what to do with the outputs. We also are not entirely sure which versions of the code to use, and were not able to find the code for a roulette algorithm. |
To find the roulette algorithm, and to learn exactly how the code that we already have works. |
| Ryan |
Found some executables for the paperclip algorithm and attempted to run them. Some worked, some did not, and we are currently still looking for any roulette algorithm and trying to figure out how to interpret the results. |
Continue searching for the Roulette algorithm and find out how to interpret the results. |
|
|
61
|
Wed Jun 3 15:02:00 2020 |
Alex Patton | Daily GENETIS Update 6/3/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Talked with Alex about results from a run of small antennas where we decreased the grid spacing. The average gain plots look different, but I don't know yet if it's definitely resolved the problem because we're still getting a decent spread on the effective volumes. Helped Eliot and Leo with a git issue and talked to Evelyn and Ryan about paperclip. They were able to run and made some images, which is a good first step. I think they're going to start on making roulette an option since paperclip only uses tournament.
Julie gave me some more details about using Karoo with ANITA data.
|
I'll try installing Karoo and getting the old libraries that it needs. I should also try getting root to work on my personal space on OSC since I'll need that for the ANITA data (which I'm also going to need to find). I have a link to a dropbox with Oindree's stuff on blastfamy so I'll try looking there for hints. |
|
Alex P.
|
Ran average gain on smaller grid spacing for small antennas and did see pattern closer to larger antenna rather than the small antenna with larger grid spacing, also set up AraSim to print out HGain which should be the effective heights, which should run the next iteration of the loop. |
Compare HGain to find how th effective heights compare and make sure they do in fact differ between runs, also continue to see how grid spacing affects the small antenna and possibly run on a previous version which gave a zero vEff |
| Eliot & Leo |
Forgot to do updates this week so this is entire week. We have the loop running with two chromosomes up through Part C. ie we edited xmacros and other related parts to run simulation/XF with an asym bicone. Part D shouldn't need any changes, and Part E/F will need some.
|
Friday/through the weekend we will each do git on codecademy and next week we will implement changes in the fitness function and plottinf(Part E/F)
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
63
|
Fri Jun 5 14:29:56 2020 |
Alex Patton | Daily GENETIS Update 6/5/20 |
| Name |
Today's Update |
Plans for Monday |
| Alex M. |
Helped Alex P try to figure out how to get data Amy was asking us to get for comparing antennas in AraSim. We've been stuck trying to figure out where all of the data is printed (there are a bunch of .cc files and quantities we aren't familiar with--for example, vmmhz in Report.cc sounds like the V/Mhz with the effective heights folded in, but we aren't sure).
I worked on some things in the paper that were mentioned in the minutes from last week's meeting, but I didn't see Amy's comments til the GENETIS meeting.
|
Monday morning Amy might stop by to talk to Alex and I about AraSim so we can try to find some of the info we haven't been able to print out. We'll also watch out for anything she posts in slack about it.
We'll present an outline of the paper at 12 on monday to the Ara group meeting to see if it needs to be an ARA paper or not.
I'm also going to keep looking for how to use the automatic grid spacing (and I'm planning on enlisting Eliot and Leo's help in being able to look at models in XF to make sure XF is designing antennas correctly).
|
|
Alex P.
|
Worked with Alex to find the volts/MHz after folding in the effective heights. While doing this we noticed a possible bug in AraSim. Currently our settings have a blank value for SIMULATION_MODE and a documentation we found says the default should be 0 but inside of AraSim the default is actually 1. Now Report.cc only uses volts/MHz if the SIMULATION MODE is set equal to 0 so this could've been an oversight when making the setup file if someone assumed leaving it blank would be zero. |
After checking whether the simulation mode is intended or an oversight, we want to check on the Volts/MHz and then also work on automatically setting the grid spacing and possibly work on implementing a penalty in the fitness score for antennas that get too small. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
64
|
Mon Jun 8 15:16:16 2020 |
Alex Patton | Daily GENETIS Update 6/8/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Worked with Alex on finding the variables Amy suggested to us in AraSim. We found the Efield and max peak voltage variables and printed them, comparing three antennas: a small one with normal veffective (nonzero), a small on with 0 veffective, and the actual bicone. The electic field was the same for all antennas, but the peak voltages were all very low for the antenna with 0 veffective. Actually, after reinvestigating, it looks like the maximum peak voltages *were* the same between all the antennas.
I've been looking into using the automatic grid spacing in XF. I think I need to get a better understanding of XF, so I started playing with it to design things in the GUI. I want to be able to make a model using the xmacros that I can look at in the GUI so I can understand all of the functions being used. Since we've checked AraSim pretty extensively, we think that the issue is with XF. I think the grid spacing might be allowing small antennas to appear better than they are.
|
I think to investigate if the problem is in XF we should do a run of the same antenna with different grid spacing and look at the average gain plots. A preliminary test could be to try modeling a small antenna with a very large grid size--if that gives a very high gain, then it would suggest that the issue is with grid spacing being too large for small antennas.
I think I'll also keep playing with Karoo. I've been reviewing some ML material to refamiliarize myself and see if I can write a few simple programs. I got Karoo running on OSC last week but I don't really understand the outputs (using the practice dataset).
|
|
Alex P.
|
Worked through more with AraSim looking at Amy's suggestions she sent over the weekend. Was able to print the PeakV and the Electric Fields, the Electric Fields matched and were the same which is what should have happened and the PeakV varied depending on how well it detected the event. Also the trigger threshold is already printed so we looked at that too. |
We believe that AraSim is functioning properly so next we will look more into XF, and the grid spacing. ProGrid we think is the option from what we talked with Cade but we will continue to look further into it. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
Was added as a collaborator to the paperclip repository on git hub and created a new dev branch and directory. Afterward, I started writing the new roulette algorithm. All changes have been pushed to the githhub repository. |
Continue writing the roulette algorithm and start work on the github classes on Codecademy. |
|