| |
ID |
Date |
Author |
Subject |
|
|
120
|
Mon Nov 23 18:02:40 2020 |
Ryan Debolt | Monday Updates |
| Alex M |
Kept working with Amy, Alex P, Julie, and Ben on the AraSim fix. We fixed our issue from last week but have a new one in stage 2. It looks like the issue has to do with resetting the values for V_forfft right before stage 2 (around line 963). Check here for the current version of Report.cc: /users/PAS0654/pattonalexo/EFieldProject/11_23_20 |
| Ryan |
Fixed the issue with the segmentation faults in the GA (simple fix one line changed) and worked with Kai to start creating a spreadsheet of results. Results seem to suggest that 2 tournament to 8 roulette seems to be the ideal selection ratio and the initial test seem to suggest a small amount of reproduction is ideal but more testing is needed to confirm this. |
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
122
|
Mon Nov 30 17:00:31 2020 |
Ethan Fahimi | Monday Updates |
| Alex M |
Kicked the loop back up. Helped Ethan and Parker with their projects during the working meeting. |
| Ryan |
Changed the tournament/roulette ratio, reproduction_no, and crossover_no to be read in variables in the GA to increase the quality of life when running the algorithm and to prep the code for some testing. Format for how to call the code is written at the top of the cpp program file. |
| Ethan F |
Worked with Alex M on further fixing AREA. The first generation works now, we are making minor fixes to get subsequent generations up and running. |
| |
|
| |
|
| |
|
| |
|
|
|
|
215
|
Mon May 1 05:45:02 2023 |
Alex M | Midscale PUEO Run | I've started a run of the PUEO loop to get some data for evolving the antenna width and height. We are running with 24 individuals, each with 400k neutrinos at an energy exponent of 19. Here is the output directory: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_05_01_Test_5
It's very important to note that the cross-pol data being used still comes from the data already stored in pueosim. This is because our extremely low cross polarizations were causing us to have no effective volumes. There's likely some issue in what values from XF we are choosing as our crosspols. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_05_01_Test_5' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=10 ## number of generations (after initial) to run through
NPOP=24 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=40000 ## Number of Neutrinos Thrown in AraSim
exp=19 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=6 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
PUEO=1 ## IF 1, we evolve the PUEO quad-ridged horn antenna, if 0, we evolve the Bicone
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=6 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=10 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
|
|
|
161
|
Tue Jun 7 14:14:21 2022 |
Dylan Wells | Matching Circuits Slides | Slides contatining my notes on matching circuits.
https://docs.google.com/presentation/d/1x25nhiqaW7LvPZ1pNZ5O4ZzsWZbtgqxBQ5haB9uWgQY/edit?usp=sharing |
|
|
191
|
Mon Feb 6 10:23:53 2023 |
Dylan Wells | Matching Circuit Parts | Attached is a spreadsheet with the information on the parts we need for the N=14 matching circuit board.
https://docs.google.com/spreadsheets/d/1x8dX3tNE-WSHjH_slj_EH4XsHcAnCVC05XiRBFPtIUc/edit#gid=0 |
|
|
137
|
Wed Sep 8 16:36:33 2021 |
Alex M | MODE Workshop Presentation | We presented at a workshop put on by the MODE collaboration. MODE is a collaboration dedicated to applying Automatic Differentiation to detector design. Here's the website: https://mode-collaboration.github.io/#:~:text=MODE%20(for%20Machine%2Dlearning%20Optimized,in%20space%2C%20and%20in%20nuclear
|
| Attachment 1: GENETIS_MODE_Presentation.pptx
|
|
|
142
|
Fri Feb 4 16:50:09 2022 |
Ryan Debolt | Loop Run |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- To test rank selection in main loop
- What is different about this run from the last?
- Rank Slection is being used.
- Parents.csv introduced.
- Elite is being turned off.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- 06% Reproduction
- 72% Crossver
- 22% Immigration
- 1% M_rate (unused)
- 5% sigma (unused)
- Selection methods used (% of each):
- 0% Elite
- 0% Reproduction
- 10% Tournament
- 90% Rank
- Are we using the database?
Directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_02_04_Rank |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_02_04_Rank' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=0 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=1 ## Percent of individuals selected through tournament (divided by 10)
RANK=9 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
128
|
Fri Jan 29 15:59:21 2021 |
Alex M | Loop Demonstration Video | Here's the video I made a few months ago demonstrating how to run the loop. There might be some changes that have been made since then, but they'll generally be minor so it should still be representative of what running the loop looks like. It might need to be shared with you through the Microsoft drive service OSU gives us in order to view it, so either message me on Slack or email me at machtay.1@osu.edu and I'll share the link through there.
https://drive.google.com/file/d/1yL_eH_w2hXNt7J7YOwS5xl3ZHwGyl5JR/view |
|
|
5
|
Thu Feb 21 14:07:14 2019 |
Julie Rolla | Logging into Nutau via XRDP | As mentioned in the manual, using ssh to login to Nutau causes delays; the XFdtd GUI cannot be surpress and must be forwarded via X11 forwarding, and this is extremely slow. We are currently looking into using a newly suggested option, XRDP. Information will be added as this process continues. |
|
|
236
|
Wed Aug 2 12:49:57 2023 |
Dylan Wells | Line of Best-Fit Straightened Sides Antenna | Previously, we have tried to straighten the sides of the best-evolved curved antenna in elog 229. However, there were potential issues with how closely this line resembled the curve of the antenna.
So, I attempted to create another straightened sides antenna using a linear regression model to find the best fitting line for the curve to create an antenna.
I made a Python notebook to separate the equations for each curve into 1000 discrete points. Then I ran a linear regression model to fit a curve of the points 1 - n, and a second curve of the n - 1000 points, looping from n = 2 to n = 999.
These results are from the combined output with the least squared error compared to the original.
Pictured is a plot showing the two sides of the curved bicone in red and blue, with the best fitting lines for each in black as well as a model in XF.
Results:
The antenna has a fitness score of 3.80627 with an error of 0.0759725.
This is much lower than the 5.71 of the curved antenna and 5.11 of the other attempt at straightening.
For the next attempt, we could consider constraining the endpoints to be the same as the original antenna to conserve the radius, and/or adding an extra line to fit the curve.
I've also attached a picture of what my notebook found for fitting 3 lines to the curve (not modeled or tested).
Professor Chen recommended using 3 sides and constraining the outer radii of the cones to match the original curved design.
Path to linear regression notebook: /users/PAS1960/dylanwells1629/developing/notebook.ipynb
Path to XF project: /users/PAS1960/dylanwells1629/straightened.xf |
| Attachment 1: output.png
| 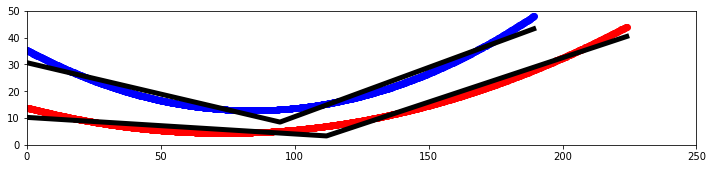 |
| Attachment 2: image_(3).png
| .png) |
| Attachment 3: output.png
| 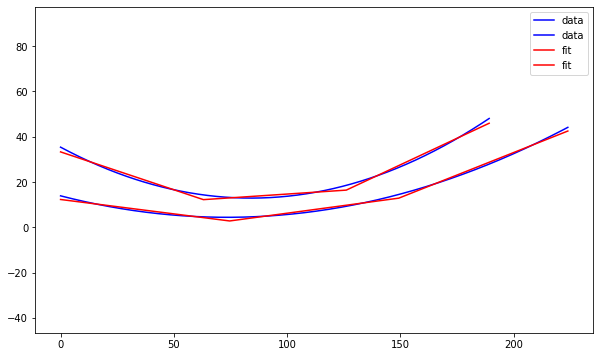 |
|
|
126
|
Tue Jan 19 19:00:30 2021 |
Alex M | Latest Run Details/Running on Slurm | OSC managed to figure out our issue with running XF from an interactive job on Slurm. Previously, we were losing our connection to x11 forwarding. The solution is to use sinteractive instead of srun to obtain an interactive job. Here's the syntax:
sinteractive -A PAS0654 -t <time> -N 1 -n 8 -p serial
The -p serial flag denotes the type of partition to request. It's important to specify this, as otherwise it will default to the debug node, which has a limit of 1 hour.
We're going to begin a new run. The title is Machtay_2021_1_15_NPOP50_Asym . It is using the latest version of the GA Ryan has been working on (Latest_GA_Asym.cpp). We're using 6% reproduciton, 72% crossover, and 22% mutation. We are using 80% roulette selection. This correlates to the three numbers passed into the GA being 3 36 8 (since we're using 50 individuals). |
|
|
179
|
Tue Aug 16 11:41:07 2022 |
Dylan Wells | Instructions for Running IceMC | Running IceMC:
Go into the directory ../anitaBuildTool/build/components/icemc/
And run the command
*May need to chmod -R 775 ../anitaBuildTool/comonents/icemc/ if you get a permissions error
inputFile:
Must be the full path to the file
Config files are found in ../anitaBuildTool/components/icemc
Ex: /users/PAS1960/dylanwells1629/anitaBuildTool/components/icemc/inputs.anita4.conf
Config files are found in ../anitaBuildTool/components/icemc
outputDirectory:
Will be made in ../anitaBuildTool/build/components/icemc/ by default, specify full path otherwise.
runNumber:
The run number.
numberOfNeutrinos:
The number of neutrinos generated in the simulation.
Can be found in inputs.conf
Default is 2,000,000.
#How many neutrinos to generate
triggerThreshold:
Threshold for each band for the trigger.
Default is 2.3
#thresholds for each band- this is only for the frequency domain voltage trigger. If using a different trigger scheme then keep these at the default values of 2.3 because the max among them is used for the chance in hell cuts
energyExponent:
The exponent of the energy for the neutrinos
Can be found in input.conf
Default is 1020
# Select energy (just enter the exponent) or (30) for baseline ES&S (1) for E^-1 (2) for E^-2 (3) for E^-3 (4) for E^-4 (5) for ES&S flux with cosmological constant (6) for neutrino GZK flux from Iron nuclei (16-22)not using spectrum but just for a single energy (101-114)use all kinds of theoretical flux models |
|
|
178
|
Wed Aug 10 22:38:20 2022 |
Dylan Wells | Instructions for Installing IceMC | Installing IceMC:
I: Getting anitaBuildTool
Clone the anitaBuildTool repository (https://github.com/anitaNeutrino/anitaBuildTool) to your user.
II: Get the Anita.sh file onto your user.
Either copy the file from /users/PAS1960/dylanwells1629/Anita.sh
or create a new file Anita.sh with the following code
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
#this modules were originally loading in both the env.sh, and bashrc_anita.sh files. This was redundant so it was added here, and removed from the others.
module load gnu/7.3.0
module load gnu
module load mvapich2
module load fftw3
#module load python/3.6-conda5.2
module load cmake
PATH=$PATH:$HOME/.local/bin:$home/bin
export PATH
export CC=`which gcc`
export CXX=`which g++`
export FFTWDIR=/fs/project/PAS0654/shared_software/fftw3/gnu/6.3/mvapich2/2.2/3.3.5
export ANITA_SOURCE_DIR=~/anitaBuildTool/
export ANITA_UTIL_INSTALL_DIR=~/anitaBuildTool/
export ICEMC_SRC_DIR=~/anitaBuildTool/components/icemc/
export ICEMC_BUILD_DIR=~/anitaBuildTool/build/components/icemc/
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
# User specific aliases and functions
#This env.sh is for running the BiconeEvolution GENETIS software. This should only be un-commented if you are running GENETIS software. When you do this, comment out env.sh.
#source ~/new_root_setup.sh
source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh
#source /cvmfs/ara.opensciencegrid.org/trunk/centos7/setup.sh
#module load python/3.6-conda5.2
#BiconeGENETIS directory shortcut SHARED
alias GE='cd ../../../fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/'
#emacs Alias
alias emacs='emacs -nw'
#root alias
alias root='root -l'
#Alias
alias l="ls"
alias python='/cvmfs/ara.opensciencegrid.org/trunk/centos7/misc_build/bin/python3.9'
Then source the file
Note: You will need access to PAS0654 for this step or you will get a permissions error.
III: Running the build tool.
Go into the anitaBuildTool directory
And run the building script
Note: There will be an error if you source files for running Ara in your .bashrc
Comment these out and restart your terminal before running the build. (remember to source Anita.sh before running the build tool. You could also source Anita.sh in your .bashrc)

Error if you source files for running Ara:
CMake Error at components/libRootFftwWrapper/cmake_install.cmake:238 (file):
file INSTALL cannot copy file
"/users/PAS1960/dylanwells1629/anitaBuildTool/components/libRootFftwWrapper/include/AnalyticSignal.h"
to
"/cvmfs/ara.opensciencegrid.org/v2.0.0/centos7/ara_build/include/AnalyticSignal.h":
Read-only file system.
Call Stack (most recent call first):
|
|
|
204
|
Mon Mar 20 16:31:09 2023 |
Alex M | Installing PUEOsim | Will sent me some info on how to install PUEOsim. I attached a markdown file he sent for how to install PUEOsim. He also sent the path to pre-compiled installation of PUEOsim. Here's the path to the pre-compiled installation: /users/PAS0654/wluszczak/PUEO_shared
Here is his full message, which details that you must also run set_env.sh before running PUEOsim:
a pre-compiled version is located at /users/PAS0654/wluszczak/PUEO_shared . Inside that directory, there's a set_env.sh script that you will need to source before running anything (hopefully this works, though there's a high probability you might run into permissions issues with the dependencies).
To run PUEOsim, use the following (see here for instructions: https://github.com/PUEOCollaboration/pueoSim):
./simulatePueo -i {inputFile} -o {outputDirectory} -r {runNumber} -n {numberOfNeutrinos} -e {energyExponent}
The outputs will be root files, but it should be possible (either modifying our own version directly or with a script) to print out the effective volume to a .txt to read.
|
| Attachment 1: CentOS_7_install.md
|
#pueoBuilder Installation Tutorial
This tutorial will guide you through the process of building the tools included in pueoBuilder from scratch, including the prerequisites and any environment variables that you will need to set. This sort of thing is always a bit of a nightmare process for me, so hopefully this guide can help you skip some of the frustration that I ran into. I did not have root acces on the system I was building on, so the instructions below are what I had to do to get things working with local installations. If you have root access, then things might be a bit easier. For reference I'm working on CentOS 7, other operating systems might have different problems that arise.
##Prerequisites
As far as I can tell, the prerequisites that need to be built first are:
-cmake 3.21.2 (I had problems with 3.11.4)
-gcc 11.1.0 (9.X will not work)
-fftw 3.3.9
-gsl 2.7.1 (for ROOT)
-ROOT 6.24.00
###CMake
You can download the source files for CMake here: https://cmake.org/download/. Untar the source files with:
tar -xzvf cmake-3.22.1.tar.gz
Compiling CMake is as easy as following the directions on the website: https://cmake.org/install/, but since we're doing a local build, we'll use the `configure` script instead of the listed `bootstrap` script. As an example, suppose that I downloaded the above tar file to `/scratch/wluszczak/cmake`:
mkdir install
cd cmake-3.22.1
./configure --prefix=/scratch/wluszczak/cmake/install
make
make install
You should additionally add this directory to your `$PATH` variable:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
To check to make sure that you are using the correct version of CMake, run:
cmake --version
and you should get:
cmake version 3.22.1
CMake suite maintained and supported by Kitware (kitware.com/cmake).
### gcc 11.1.0
Download the gcc source from github here: https://github.com/gcc-mirror/gcc/tags. I used the 11.1.0 release, though there is a more recent 11.2.0 release that I have not tried. Once you have downloaded the source files, untar the directory:
tar -xzvf gcc-releases-gcc-11.1.0.tar.gz
Then install the prerequisites for gcc:
cd gcc-releases-gcc-11.1.0
contrib/download_prerequisites
One of the guides I looked at also recommended installing flex separately, but I didn't seem to need to do this, and I'm not sure how you would go about it without root priviledges, though I imagine it's similar to the process for all the other packages here (download the source and then build by providing an installation prefix somewhere)
After you have installed the prerequisites, create a build directory:
cd ../
mkdir build
cd build
Then configure GCC for compilation like so:
../gcc-releases-gcc-11.1.0/configure -v --prefix=/home/wluszczak/gcc-11.1.0 --enable-checking=release --enable-languages=c,c++,fortran --disable-multilib --program-suffix=-11.1
I don't remember why I installed to my home directory instead of the /scratch/ directories used above. In principle the installation prefix can go wherever you have write access. Once things have configured, compile gcc with:
make -j $(nproc)
make install
Where `$(nproc)` is the number of processing threads you want to devote to compilation. More threads will run faster, but be more taxing on your computer. For reference, I used 8 threads and it took ~15 min to finish.
Once gcc is built, we need to set a few environment variables:
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
We also need to make sure cmake uses this compiler:
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/bin/gfortran-11.1
If your installation prefix in the configure command above was different, substitute that directory in place of `/home/wluszczak/gcc-11.1.0` for all the above export commands. To easily set these variables whenever you want to use gcc-11.1.0, you can stick these commands into a single shell script:
#load_gcc11.1.sh
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
(again substituting your installation prefix in place of mine). You can then set all these environment variables by simply running:
source load_gcc11.1.sh
Once this is done, you can check that gcc-11.1.0 is properly installed by running:
gcc-11.1 --version
Note that plain old
gcc --version
might still point to an older version of gcc. This is fine though.
###FFTW 3.3.9
Grab the source code for the appropriate versino of FFTW from here: http://www.fftw.org/download.html
However, do NOT follow the installation instructions on the webpage. Those instructions might work if you have root privileges, but I personally couldn't seem to to get things to work that way. Instead, we're going to build fftw with cmake. Untar the fftw source files:
tar -xzvf fftw-3.3.9.tar.gz
Make a build directory and cd into it:
mkdir build
cd build
Now build using cmake, using the flags shown below. For reference, I downloaded and untarred the source file in `/scratch/wluszczak/fftw/build`, so adjust your install prefix accordingly to point to your own build directory that you created in the previous step.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON ../fftw-3.3.9
make install -j $(nproc)
Now comes the weird part. Remove everything except the `include` and `lib64` directories in your build directory (if you installed to a different `CMAKE_INSTALL_PREFIX`, the include and lib64 directories might be located there instead. The important thing is that you want to remove everything, but leave the `include` and `lib64` directories untouched):
rm *
rm -r CMakeFiles
Now rebuild fftw, but with an additional flag:
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON -DENABLE_FLOAT=ON ../fftw-3.3.9
make install -j $(nproc)
At the end of the day, your fftw install directory should have the following files:
include/fftw3.f
include/fftw3.f03
include/fftw3.h
include/fftw3l.f03
include/fftw3q.f03
lib64/libfftw3f.so
lib64/libfftw3f_threads.so.3
lib64/libfftw3_omp.so.3.6.9
lib64/libfftw3_threads.so
lib64/libfftw3f_omp.so
lib64/libfftw3f.so.3
lib64/libfftw3f_threads.so.3.6.9
lib64/libfftw3.so
lib64/libfftw3_threads.so.3
lib64/libfftw3f_omp.so.3
lib64/libfftw3f.so.3.6.9
lib64/libfftw3_omp.so
lib64/libfftw3.so.3
lib64/libfftw3_threads.so.3.6.9
lib64/libfftw3f_omp.so.3.6.9
lib64/libfftw3f_threads.so
lib64/libfftw3_omp.so.3
lib64/libfftw3.so.3.6.9
Why do we have to do things this way? I don't know, I'm bad at computers. Maybe someone more knowledgeable knows. I found that when I didn't do this step, I'd run into errors that pueoBuilder could not find some subset of the required files (either the ones added by building with `-DENABLE_FLOAT`, or the ones added by building without `-DENABLE_FLOAT`).
Once fftw has been installed, export your install directory (the one with the include and lib64 folders) to the following environment variable:
export FFTWDIR=/scratch/wluszczak/fftw/build
Again, substituting your own fftw install prefix that you used above in place of `/scratch/wluszczak/fftw/build`
###gsl 2.7.1
gsl 2.7.1 is needed for the `mathmore` option in ROOT. If you have an outdated version of gsl, ROOT will still compile, but it will skip installing `mathmore` and `root-config --has-mathmore` will return `no`. To fix this, grab the latest source code for gsl from here: https://www.gnu.org/software/gsl/. Untar the files to a directory of your choosing:
tar -xzvf gsl-latest.tar.gz
For some reason I also installed gsl to my home directory, but in principle you can put it wherever you want.
mkdir /home/wluszczak/gsl
./configure --prefix=/home/wluszczak/gsl
make
make check
make install
To make sure ROOT can find this installation of gsl, you'll again need to set an environment variable prior to building ROOT:
export GSL_ROOT_DIR=/home/wluszczak/gsl/
I also added this to my $PATH variable, though I don't remember if that was required to get things working or not:
export PATH=/home/wluszczak/gsl/bin/:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gsl/lib:$LD_LIBRARY_PATH
###ROOT 6.24.00
Download the specific version of ROOT that you need from here: https://root.cern/install/all_releases/
You might need to additionally install some of the dependencies (https://root.cern/install/dependencies/), but it seems like everything I needed was already installed on my system.
Untar the source you downloaded:
tar -xzvf root_v6.24.00.source.tar.gz
Make some build and install directories:
mkdir build install
cd build
Run CMake, but be sure to enable the fortan, mathmore and minuit2 options. For reference, I had downloaded and untarred the source files to `/scratch/wluszczak/root`. Your installation and source paths will be different.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/root/install/ /scratch/wluszczak/root/root-6.24.00/ -Dfortran=ON -Dminuit2=ON -Dmathmore=ON
Note: if you end up with an error related to compiling XROOTD, then add -Dxrootd=OFF to the original cmake command above.
Then proceed to start the build:
cmake --build . --target install -j $(nproc)
If everything has worked then after the above command finishes running, you should be able to run the following file to finish setting up ROOT:
source ../install/bin/thisroot.sh
##pueoBuilder
By this point, you should have working installations of CMake 3.21.2, gcc-11.1.0, fftw 3.3.9, and ROOT 6.24.00. Additionally, the following environment variables should have been set:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
export FFTWDIR=/scratch/wluszczak/fftw/build
At this point, the hard work is mostly done. Check out pueoBuilder with:
git clone git@github.com:PUEOCollaboration/pueoBuilder
set the following environment variables:
export PUEO_BUILD_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export PUEO_UTIL_INSTALL_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export NICEMC_SRC=${PUEO_BUILD_DIR}/components/nicemc
export NICEMC_BUILD=${PUEO_BUILD_DIR}/build/components/nicemc
export PUEOSIM_SRC=${PUEO_BUILD_DIR}/components/pueoSim
export LD_LIBRARY_PATH=${PUEO_UTIL_INSTALL_DIR}/lib:$LD_LIBRARY_PATH
Where $PUEO_BUILD_DIR and $PUEO_UTIL_INSTALL_DIR point to where you cloned pueoBuilder to (in my case, `/scratch/wluszczak/PUEO/pueoBuilder`. Now you should be able to just run:
./pueoBuilder.sh
Perform a prayer to the C++ gods while you're waiting for it to compile, and hopefully at the end of the day you'll have a working set of PUEO software.
##Issues I Ran Into
If you already have an existing installation of ROOT, you may still need to recompile to make sure you're using the same c++ standard that the PUEO software is using. I believe the pre-compiled ROOT binaries available through their website are insufficient, though maybe someone else has been able to get those working.
If you're running into errors about c++ standard or compiler version even after you have installed gcc-11.1.0, then for some reason your system isn't recognizing your local installation of gcc-11.1.0. Check the path variables ($PATH and $LD_LIBRARY_PATH) to make sure the gcc-11.1.0 `bin` directory is being searched.
If you're running into an error that looks like:
CMake Error: The following variables are used in this project, but they are set to NOTFOUND.
Please set them or make sure they are set and tested correctly in the CMake files:
FFTWF_LIB (ADVANCED)
then pueoBuilder can't seem to find your fftw installation (or files that are supposed to be included in that installation), try rebuilding with different flags according to which files it seems to think are missing.
If it seems like pueoBuilder can't seem to find your fftw installation at all (i.e. you're getting some error that looks like `missing: FFTW_LIBRARIES` or `missing: FFTW_INCLUDES`), check the environment variables that are supposed to point to your fftw installation (`$FFTWDIR`) and make sure there are the correct files in the `lib` and `include` subdirectories.
|
|
|
157
|
Mon Jun 6 14:19:59 2022 |
Alex M | Important Runs (2) | This is a duplicate post of a previous post from the end of 2020 where I listed the important runs with some of their details in a table (as below). I am extending this table to include the important runs that have been conducted since this post. This includes the run used for the paper as well as the curved run done earlier this year.
In order to access the data for these runs, you can find them by going to this directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs
Some of these runs can also be accessed in the old project space directory, though they should all be contained in the above directory. Here's the path if interested:
The runs are contained in directories available in the above path. Use caution when listing files in some of these directories--some contain many files (primarily the .uan files -- more recent runs are better organized), which means it may take a long time to list files/directories.
| Name |
Description |
Symmetry |
NPOP |
Generations |
Roulette/Tourney/Rank |
Crossover |
Reproduction |
Mutation |
Injection |
Penalty |
Neutrinos |
|
| Machtay_20200824_Real_Run |
First real run with significant amounts of data after the summer improvements.______________________________________
|
Symmetric |
10 |
15 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
100k |
|
| Machtay_20200827_Asym_Length_Run |
First asymmetric length run after summer improvements. |
Asymmetric length |
10 |
17 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
100k |
|
| Machtay_20200831_Asym_Length_and_Angle |
Asymmetric length and angle run after summer improvements. |
Asymmetric length and angle |
10 |
42 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
150k |
|
| Machtay_20200911_Symmetric |
Longer symmetric run with fewer neutrinos. |
Symmetric |
10 |
35 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
30k |
|
| Machtay_20200914_Asymmetric_50_Individuals |
Longer asymmetric run with fewer neutrinos. |
Asymmetric (all dimensions) |
50 |
26 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
30k |
|
| Machtay_20201016_Symmetric_Improved_GA |
First run using improvements to GA based on Ryan's paperclip/fast loop analysis. |
Symmetric |
50 |
10 |
50/50/0 |
75% |
10% |
- |
15% |
Yes |
30k |
|
| Machtay_20201023_300K_Nus_50_Individuals |
Started with all identical individuals to demonstrate evolution; replaced penalty with hard cutoff. Increased Nus for higher fitness score precision. |
Asymmetric (all dimensions |
50 |
25 |
50/50/0 |
75% |
10% |
- |
15% |
No |
300k |
|
| AraSim_Polarity_Fix_2021_03_19 |
Run used in the paper. In this run, we fixed an error that had been noticed by Brian and Jorge in AraSim. The error involved the polarity of the signals in Report.cc (hence the name of this run). |
Asymmetric(all dimensions) |
50 |
31 |
80/20/0 |
72% |
6% |
- |
22% |
No |
300k |
|
| 2022_02_08_Rank_Test |
This was the first long run done using a new gene for the curvature of the cones. We recast the side lengths to be described by the coefficients of a quadratic polynomial, rather than by the opening angle. This also used rank selection instead of roulette.
Additionally, mutation has been changed here so as to apply small perturbations to existing genes rather than regenerating those genes altogether. This only applies to individuals created by crossover. The mutation column indicates the probability of mutating a gene and the standard deviation of the gaussian that determines the change (in terms of % of the original value).
|
Asymmetric, Quadratic |
50 |
50 |
0/90/10 |
72% |
6% |
1%, 5% |
22% |
No |
300k |
|
| 2022_04_14_Identical_Asym_Lower_Min |
This run used the asymmetric GA to see if by lowering the minimum length (down to 10 cm instead of 37.5) the GA would try to run away to ever smaller lengths. |
Asymmetric |
50 |
6 |
2/8/0 |
72% |
6% |
- |
22% |
No |
300k |
|
|
|
|
124
|
Mon Dec 7 22:51:08 2020 |
Alex M | Important Runs | Today I removed some of the run directories which had very little or no data or weren't worth keeping around. There are still a few that I think can be removed, but I'm keeping them until we can get a consensus that they can definitely be removed. Below I listed the names and descriptions of the runs that I think we should definitely preserve going forward. In general, the more data contained in the run directory, the more important it is to keep around.
| Name |
Description |
Symmetry |
NPOP |
Generations |
Roulette/Tourney |
Crossover |
Reproduction |
Mutation |
Penalty |
Neutrinos |
|
| Machtay_20200824_Real_Run |
First real run with significant amounts of data after the summer improvements.______________________________________
|
Symmetric |
10 |
15 |
100% Roulette |
100% |
0% |
100% |
Yes |
100k |
|
| Machtay_20200827_Asym_Length_Run |
First asymmetric length run after summer improvements. |
Asymmetric length |
10 |
17 |
100% Roulette |
100% |
0% |
100% |
Yes |
100k |
|
| Machtay_20200831_Asym_Length_and_Angle |
Asymmetric length and angle run after summer improvements. |
Asymmetric length and angle |
10 |
42 |
100% Roulette |
100% |
0% |
100% |
Yes |
150k |
|
| Machtay_20200911_Symmetric |
Longer symmetric run with fewer neutrinos. |
Symmetric |
10 |
35 |
100% Roulette |
100% |
0% |
100% |
Yes |
30k |
|
| Machtay_20200914_Asymmetric_50_Individuals |
Longer asymmetric run with fewer neutrinos. |
Asymmetric (all dimensions) |
50 |
26 |
100% Roulette |
100% |
0% |
100% |
Yes |
30k |
|
| Machtay_20201016_Symmetric_Improved_GA |
First run using improvements to GA based on Ryan's paperclip/fast loop analysis. |
Symmetric |
50 |
10 |
50/50 |
75% |
10% |
15% |
Yes |
30k |
|
| Machtay_20201023_300K_Nus_50_Individuals |
Started with all identical individuals to demonstrate evolution; replaced penalty with hard cutoff. Increased Nus for higher fitness score precision. |
Asymmetric (all dimensions |
50 |
25 |
50/50 |
75% |
10% |
15% |
No |
300k |
|
| |
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
152
|
Fri Apr 8 16:07:22 2022 |
Alex M | Identical Asymmetric Lowered Length Minimum Run | We are trying to do a short run of the asymmetric bicone so that we can see how it tries to evolve the antenna when the minimum length is lowered from 37.5 cm to 10 cm. Currently, there is a problem with the loop in the asymmetric version. Attached is the run detail file. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_04_07_Identical_Asym' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=0 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=8 ## Percent of individuals selected through tournament (divided by 10)
RANK=0 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
184
|
Mon Oct 24 17:44:51 2022 |
Ryan Debolt | Icemc inputs | Here is our current assumption of the antenna angles needed for the icemc inputs. |
| Attachment 1: 261D6FA3-5612-412F-80FA-8AD961F7331A.jpeg
| 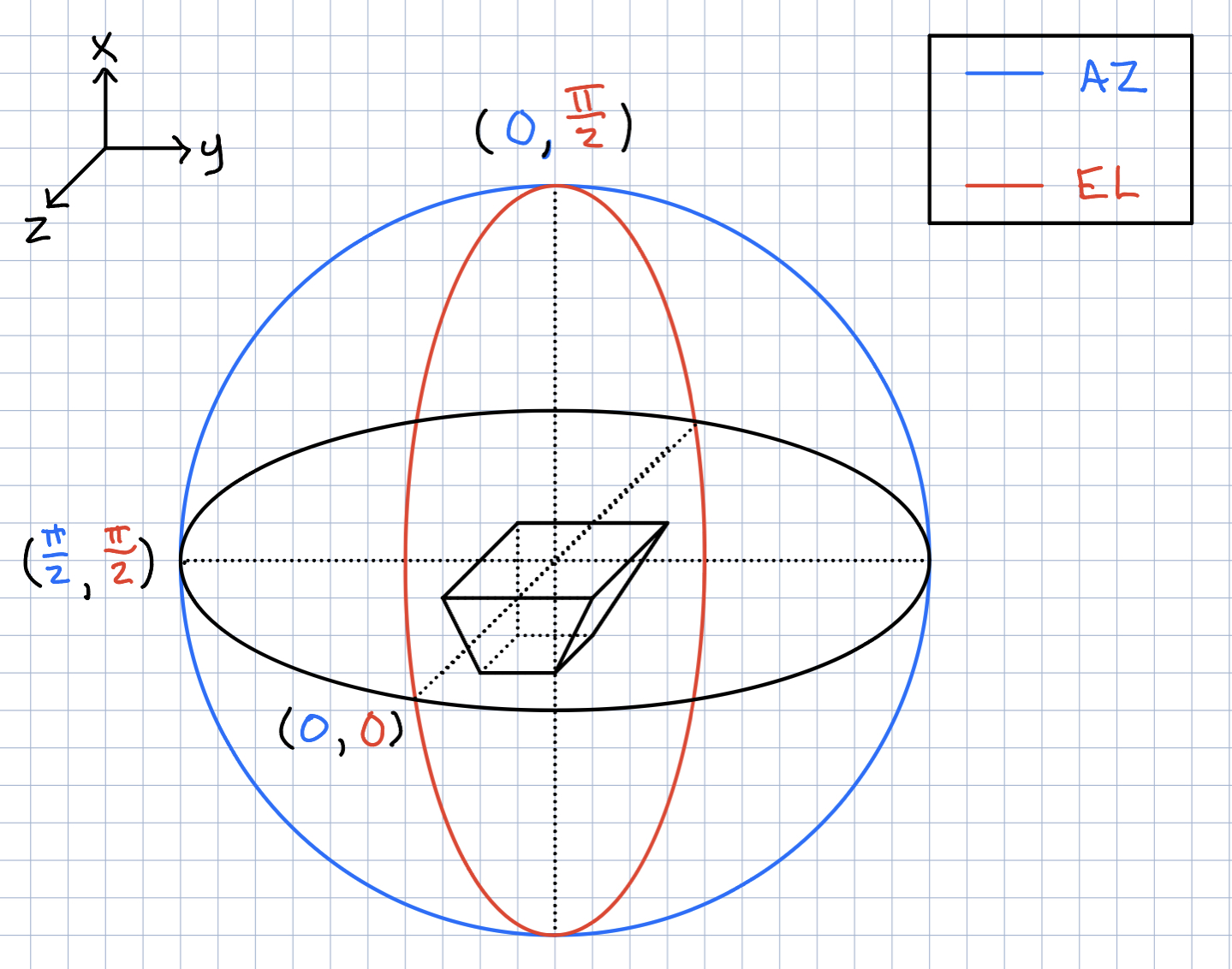 |
|
|
182
|
Fri Sep 2 12:40:17 2022 |
Alan S | Ice Shell Around Bicone | Here is a PDF with a summary/log of my work trying to integrate an ice shell surrounding the bicone antennas in the GENETIS loop. I made gain patterns of the ARA bicone with the ice shells and without them, showing the impact of the ice on the antenna's gain. Having an ice shell changes these patterns, which are also dependent on the radius of the shell. I showed that convergence on the gain patterns and expected features start ocurring for shells with radii larger than 10 meters. I also keep track of the computation time which increases with bigger shells.
Note: I also add PowerPoint slides that contain all the graphs that I made. The PDF does not contain some of those since I didn't think they were coherent with my write-up, but I may be making an omission.
|
| Attachment 1: XF_ARA_bicone_in_ice_and_air_column.pptx
|
| Attachment 2: Far_Zone_Gain_of_ARA_Bicone_insice_Ice.pdf
|
|
|
217
|
Wed May 10 17:43:20 2023 |
Alex M | How to run the loop | In this post, I'm going to give step by step instructions on everything you need to do to run the loop. See Julie's thesis for the most recent iteration of this manual. Also refer to entry 123 for a list of some errors we have encountered historically in the loop and how to resolve them, though some fixes may be outdated.
Running the loop
Getting started
Assuming you have followed the instructions from the onboarding tutorials, you should be ready to access OSC through the interactive website. Instead of using ssh to access OSC, the loop requires that you go to this website: https://ondemand.osc.edu/ . Login with your usual OSC credentials. Once there, you will need to request a Lightweight Desktop by clicking on "Interactive Apps" at the top of the page. See the screenshots attached for how to do this. Old documentation may refer to the Lightweight Desktop as a VDI.
You can request up to 24 hours for a Lightweight Desktop, and it should begin almost immediately. After you request it, the website will take you directly to the waiting page. You can navigate to this page yourself by clicking "My Interactive Sessions" at the top of the ondemand page. When the job begins, you will see a blue buttong that says "Launch Lightweight Desktop". Click it and you should have a new tab open that brings you to a remote desktop environment on OSC that you interact with through your browser.
New runs
If you're starting a new run for the first time (that is, beginning from generation 0), you will need to edit the script that runs the loop. Here is the path: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Scripts/Asym_XF_Loop.sh . At the top of the script is a list of variables that can be changed. See the first screenshot attached here.
For a new run, you must change the variable <RunName>, which is the first variable in the list. It should be in quotes and the convention is to start the name off with the date of the run, using YYYY/MM/DD format--this will always keep everything in chronological order when listing directories with <ls>. Other variables are explained in the comments next to them in Asym_XF_Loop.sh and you should change them depending on the type of run you are conducting.
To begin the loop, type in the command ./Loop_Scripts/Asym_XF_Loop.sh from the Evolutionary_Loop directory.
When the loop begins, you will be asked to press any key. After you hit a key, the loop will run Part A, which executes the GA using the <start> mode. It will then automatically move on to Part B, which creates the antennas in XFdtd. XF will ask you if you want to specify a location for the XF file to be saved. Click <No>. The loop will automatically save the XF directory into the run directory when it finishes with the first part of part B. In principle (that is, barring errors that stall the loop or interruptions due to the VDI job ending), you will never need to manually enter anything to the loop for it to run beyond this point.
However, if it is your first time using XF, you will need to set the max GHz setting upon startup to 2 instead of the default 10.
Continuing Runs
After starting a new run, the loop will run automatically. When your Lightweight Desktop job ends, you'll need to get another one repeat the steps above. You will not be prompted for any input.
Stepping Back
At times, you may want to continue the loop from an earlier point or interrupt the loop and step back. Usually you'll only want to do this within a generation, but it's possible that you will encounter an error or bug at the end of a generation and wish to return back to that point to remedy it before continuing forward in the next generation. This is a very tricky and delicate process that requires a lot of attention to detail. In the future, we'd like to make an option for the loop to do this automatically when given input at the beginning, but implementing that will take a long time and will be very sophisticated.
The state of the loop is recorded in the savestate file located here: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/saveStates . The file will be named <Run_Name>.savestate.txt . In it, you will see three numbers. The first one represents the current generation of the loop. The second one represents the state of the loop, that is, which part in the generation it is currently on. The third one represents the individual it is processing. In practice, the third number is rarely relevant, and is almost always 1, though it can be used for Part B, where the loop is processing individuals sequentially, so if it was interrupted on individual eight (for example) you can change the number to be 8 so that it will pick up where it left off without needing to redo the first seven individuals.
If you want to step back to an earlier state in a generation, you'll need to interrupt the loop and revert the state by changing the second number in the savestate file. This is NOT all you will have to do, however. Because of the way files are edited and moved around during states, you will need to manually revert files to be in the format or location needed by the earlier state.
For example, you may need to revert from state 2 (when XF is creating the antenna geometries) to state 1 (when the GA generates new individuals). In this case, you'll need to edit the savestate file to change the second number from a 2 to a 1. Then, you'll need to change the generationDNA.csv file in Generation_Data to use the values from the previous generation, since the GA will have overwritten that file and uses it to determine what individuals are to be selected from. To do this, you'll need to locate the <gen>_generationDNA.csv file from the previous generation in the <RunName> directory in Run_Outputs. This is should be found here: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/<RunName>/Generation_Data .
The above is a relatively simple example of how to step back in the loop. Stepping back from a late state to an early state will be more complicated, though if you're willing to sacrifice run time you can pretty easily follow the above to just restart a generation. Stepping back to a previous generation will be explained in a future update to this post. |
| Attachment 1: Ondemand_Front_Page.png
|  |
| Attachment 2: Ondemand_Interactive_Apps.png
| 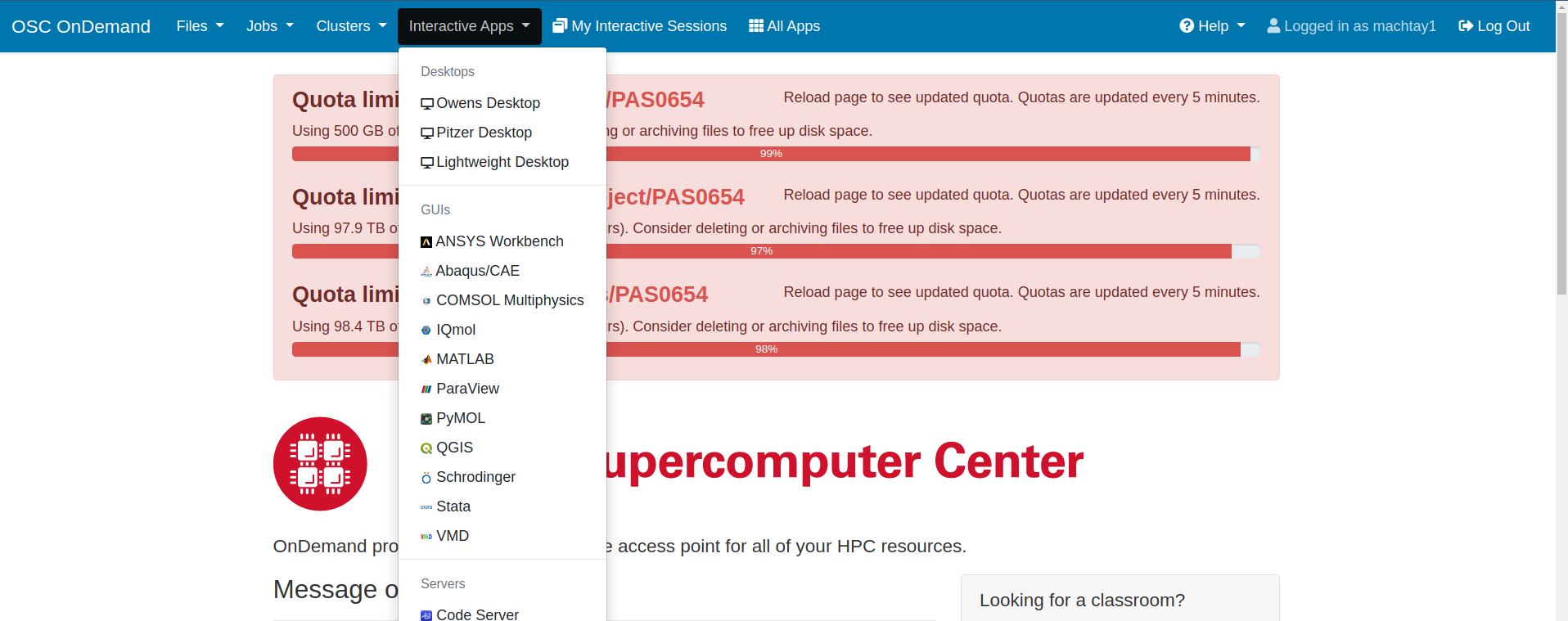 |
| Attachment 3: Launching_Lightweight_Desktop.png
| 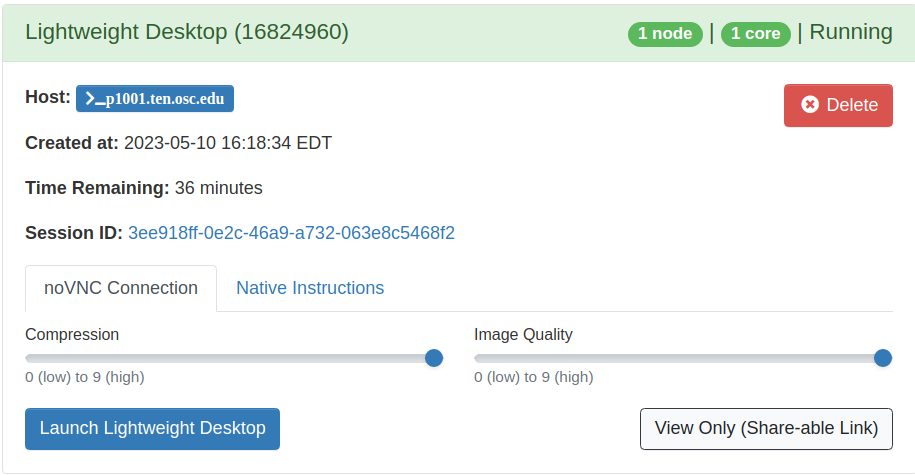 |
|