| |
ID |
Date |
Author |
Subject |
|
|
237
|
Wed Aug 2 22:33:18 2023 |
Amy | slides from building meeting 8/2/23 | Slides shown at experts building meeting Aug. 2nd, 2023. |
| Attachment 1: GENETIS_building_080223.pdf
|
|
|
180
|
Thu Aug 18 10:32:17 2022 |
Amy | reading about HPol antennas | Here are some reading materials about HPol antennas:
This paper discusses some initial HPol designs that were tested in a prototype station of ARA. We went with the slotted cylinder design.
Here you can see what the RNO-G experiment in Greenland has considered.
We can add other sources here as we find them. |
|
|
195
|
Wed Feb 15 16:33:17 2023 |
Jacob Weiler | pueoSim Input Files | Input File Format for pueoSim (Also ICEMC)
Frequency Range: From 200 MHz to 1500 MHz incrementing by 10 MHz steps
There are 8 different files that are required for pueoSim. They are:
vv_0: Max Gain at each Frequency, Vertical Polarization
hh_0: Max Gain at each Frequency, Horizontal Polarization
vh_0: Max Gain at each Frequency, Vertical to Horizontal Polarization
hv_0: Max Gain at each Frequency, Horizontal to Vertical Polarization
vv_el: Gain at Theta Angles [5, 10, 20, 30, 45, 90] for each Frequency, Vertical Polarization
vv_az: Gain at Phi Angles [5, 10, 20, 30, 45, 90] for each Frequency, Vertical Polarization
hh_el: Gain at Theta Angles [5, 10, 20, 30, 45, 90] for each Frequency, Horizontal Polarization
hh_az: Gain at Phi Angles [5, 10, 20, 30, 45, 90] for each Frequency, Horizontal Polarization
Currently XF doesn't output all the information we need to create all these files. The only files able to be made from current XF outputs are vv_0, vv_el, and vv_az. Once we have the correct XF Outputs, it shouldn't be too much of a hassle to fix the current translation code.
Format for each of the files are the same. One column is Frequency (Hz) and the other is Gain. Attached are example files vv_0 and vv_el to visually see the format, though the frequency range is the current ARA range and not the final PUEO range as I am using ARA data to make these files. The files are named vv_0 (no .txt or .csv or any extension) and vv_el
Link to Google Doc with this information: https://docs.google.com/document/d/1iRUF6hIEyQfMK0LL21caRuHPgXYP30ZdkSkFv_-Y8R0/edit |
| Attachment 1: vv_0_1
|
8.33e+07 2.11822
1.00e+08 0.9436538
1.17e+08 1.205556
1.33e+08 2.149637
1.50e+08 2.729287
1.67e+08 2.619639
1.83e+08 2.532374
2.00e+08 2.289813
2.17e+08 2.406259
2.33e+08 2.442176
2.50e+08 2.628373
2.67e+08 2.359219
2.83e+08 1.761219
3.00e+08 1.502431
3.17e+08 2.106109
3.33e+08 2.71283
3.50e+08 3.247887
3.67e+08 3.524175
3.83e+08 3.615199
4.00e+08 3.448137
4.17e+08 3.003371
4.33e+08 2.217343
4.50e+08 0.9601937
4.67e+08 -0.757537
4.83e+08 -3.06678
5.00e+08 -5.635237
5.17e+08 -8.2271
5.33e+08 -9.618924
5.50e+08 -9.791239
5.67e+08 -9.201792
5.83e+08 -9.086395
6.00e+08 -9.560636
6.17e+08 -10.94548
6.33e+08 -13.11583
6.50e+08 -16.3255
6.67e+08 -22.45401
6.83e+08 -34.91041
7.00e+08 -18.52293
7.17e+08 -12.31584
7.33e+08 -8.076791
7.50e+08 -5.287256
7.67e+08 -3.399354
7.83e+08 -2.394224
8.00e+08 -2.031315
8.17e+08 -2.07204
8.33e+08 -2.531995
8.50e+08 -3.185455
8.67e+08 -4.223504
8.83e+08 -5.275337
9.00e+08 -6.274862
9.17e+08 -6.532893
9.33e+08 -6.242696
9.50e+08 -5.514515
9.67e+08 -4.831669
9.83e+08 -4.240883
1.00e+09 -3.779134
1.02e+09 -3.558248
1.03e+09 -3.38254
1.05e+09 -3.543484
1.07e+09 -3.611961
|
| Attachment 2: vv_el_1
|
8.33e+07 -19.61153
1.00e+08 -21.05896
1.17e+08 -21.14054
1.33e+08 -20.63561
1.50e+08 -20.56293
1.67e+08 -21.11712
1.83e+08 -21.6924
2.00e+08 -22.23107
2.17e+08 -22.38623
2.33e+08 -22.01034
2.50e+08 -20.95566
2.67e+08 -19.49215
2.83e+08 -19.23056
3.00e+08 -21.02166
3.17e+08 -24.84512
3.33e+08 -30.42641
3.50e+08 -30.39998
3.67e+08 -24.79053
3.83e+08 -20.92692
4.00e+08 -18.28975
4.17e+08 -16.42205
4.33e+08 -15.10437
4.50e+08 -14.27861
4.67e+08 -13.84517
4.83e+08 -13.72481
5.00e+08 -13.71555
5.17e+08 -13.67698
5.33e+08 -13.47088
5.50e+08 -13.12691
5.67e+08 -12.80078
5.83e+08 -12.76288
6.00e+08 -13.29937
6.17e+08 -14.53883
6.33e+08 -16.51551
6.50e+08 -19.31712
6.67e+08 -23.38054
6.83e+08 -28.77149
7.00e+08 -26.26056
7.17e+08 -20.68733
7.33e+08 -16.92428
7.50e+08 -14.40974
7.67e+08 -12.83548
7.83e+08 -11.95044
8.00e+08 -11.46287
8.17e+08 -11.11874
8.33e+08 -10.75576
8.50e+08 -10.35075
8.67e+08 -9.963577
8.83e+08 -9.712427
9.00e+08 -9.691508
9.17e+08 -9.958833
9.33e+08 -10.48243
9.50e+08 -11.25391
9.67e+08 -12.16331
9.83e+08 -13.22009
1.00e+09 -14.23114
1.02e+09 -15.10789
1.03e+09 -15.45052
1.05e+09 -15.20106
1.07e+09 -14.44125
8.33e+07 -13.61057
1.00e+08 -15.05177
1.17e+08 -15.12521
1.33e+08 -14.60951
1.50e+08 -14.52322
1.67e+08 -15.06334
1.83e+08 -15.62302
2.00e+08 -16.1491
2.17e+08 -16.29245
2.33e+08 -15.91477
2.50e+08 -14.8618
2.67e+08 -13.39692
2.83e+08 -13.1095
3.00e+08 -14.84329
3.17e+08 -18.54948
3.33e+08 -24.02788
3.50e+08 -24.85216
3.67e+08 -19.17134
3.83e+08 -15.1775
4.00e+08 -12.46181
4.17e+08 -10.53846
4.33e+08 -9.176889
4.50e+08 -8.313917
4.67e+08 -7.849362
4.83e+08 -7.705056
5.00e+08 -7.681289
5.17e+08 -7.638247
5.33e+08 -7.434054
5.50e+08 -7.090922
5.67e+08 -6.756147
5.83e+08 -6.694556
6.00e+08 -7.188318
6.17e+08 -8.361327
6.33e+08 -10.23697
6.50e+08 -12.87492
6.67e+08 -16.64803
6.83e+08 -21.92072
7.00e+08 -21.02616
7.17e+08 -15.29548
7.33e+08 -11.32251
7.50e+08 -8.680712
7.67e+08 -7.026238
7.83e+08 -6.092156
8.00e+08 -5.577537
8.17e+08 -5.222755
8.33e+08 -4.855792
8.50e+08 -4.445815
8.67e+08 -4.04485
8.83e+08 -3.769133
9.00e+08 -3.712655
9.17e+08 -3.934655
9.33e+08 -4.405427
9.50e+08 -5.116381
9.67e+08 -5.962245
9.83e+08 -6.955142
1.00e+09 -7.921253
1.02e+09 -8.79726
1.03e+09 -9.21963
1.05e+09 -9.101768
1.07e+09 -8.46457
8.33e+07 -7.672357
1.00e+08 -9.089356
1.17e+08 -9.131313
1.33e+08 -8.574198
1.50e+08 -8.436004
1.67e+08 -8.923278
1.83e+08 -9.424584
2.00e+08 -9.903976
2.17e+08 -10.00275
2.33e+08 -9.618365
2.50e+08 -8.573076
2.67e+08 -7.108304
2.83e+08 -6.730207
3.00e+08 -8.258189
3.17e+08 -11.54483
3.33e+08 -16.48101
3.50e+08 -20.44362
3.67e+08 -14.99668
3.83e+08 -10.39325
4.00e+08 -7.322492
4.17e+08 -5.15867
4.33e+08 -3.614434
4.50e+08 -2.600771
4.67e+08 -2.012469
4.83e+08 -1.773685
5.00e+08 -1.692136
5.17e+08 -1.631185
5.33e+08 -1.436829
5.50e+08 -1.104481
5.67e+08 -0.7477778
5.83e+08 -0.6082266
6.00e+08 -0.9526983
6.17e+08 -1.891183
6.33e+08 -3.416407
6.50e+08 -5.508213
6.67e+08 -8.350483
6.83e+08 -12.36274
7.00e+08 -15.94441
7.17e+08 -11.94184
7.33e+08 -7.215725
7.50e+08 -4.031596
7.67e+08 -2.033106
7.83e+08 -0.8896422
8.00e+08 -0.2599349
8.17e+08 0.1364451
8.33e+08 0.508579
8.50e+08 0.9189453
8.67e+08 1.351561
8.83e+08 1.703014
9.00e+08 1.881713
9.17e+08 1.823472
9.33e+08 1.546895
9.50e+08 1.061567
9.67e+08 0.4561304
9.83e+08 -0.2820854
1.00e+09 -1.030102
1.02e+09 -1.782719
1.03e+09 -2.307445
1.05e+09 -2.516682
1.07e+09 -2.306051
8.33e+07 -4.297208
1.00e+08 -5.677257
1.17e+08 -5.671377
1.33e+08 -5.051709
1.50e+08 -4.836023
1.67e+08 -5.245572
1.83e+08 -5.661229
2.00e+08 -6.072461
2.17e+08 -6.104968
2.33e+08 -5.710936
2.50e+08 -4.682109
2.67e+08 -3.235083
2.83e+08 -2.745363
3.00e+08 -3.999428
3.17e+08 -6.728222
3.33e+08 -10.71871
3.50e+08 -16.85379
3.67e+08 -15.67102
3.83e+08 -9.610837
4.00e+08 -5.713407
4.17e+08 -3.051436
4.33e+08 -1.161082
4.50e+08 0.1202912
4.67e+08 0.9189369
4.83e+08 1.315152
5.00e+08 1.493735
5.17e+08 1.58528
5.33e+08 1.756137
5.50e+08 2.045786
5.67e+08 2.395989
5.83e+08 2.609925
6.00e+08 2.448707
6.17e+08 1.814713
6.33e+08 0.7368893
6.50e+08 -0.702895
6.67e+08 -2.543868
6.83e+08 -4.926394
7.00e+08 -7.929486
7.17e+08 -9.722289
7.33e+08 -6.876765
7.50e+08 -3.230588
7.67e+08 -0.6956752
7.83e+08 0.8078526
8.00e+08 1.641694
8.17e+08 2.100993
8.33e+08 2.439688
8.50e+08 2.768471
8.67e+08 3.145787
8.83e+08 3.516955
9.00e+08 3.809434
9.17e+08 3.95456
9.33e+08 3.942741
9.50e+08 3.779913
9.67e+08 3.52297
9.83e+08 3.16981
1.00e+09 2.79626
1.02e+09 2.387999
1.03e+09 2.060452
1.05e+09 1.836171
1.07e+09 1.800864
8.33e+07 -1.144795
1.00e+08 -2.455938
1.17e+08 -2.361629
1.33e+08 -1.627502
1.50e+08 -1.273074
1.67e+08 -1.547562
1.83e+08 -1.814825
2.00e+08 -2.110694
2.17e+08 -2.028768
2.33e+08 -1.626653
2.50e+08 -0.6542816
2.67e+08 0.689521
2.83e+08 1.297091
3.00e+08 0.4526482
3.17e+08 -1.408842
3.33e+08 -3.855139
3.50e+08 -7.45269
3.67e+08 -13.8242
3.83e+08 -21.86369
4.00e+08 -10.18875
4.17e+08 -4.926253
4.33e+08 -1.717269
4.50e+08 0.408406
4.67e+08 1.793992
4.83e+08 2.602153
5.00e+08 3.033494
5.17e+08 3.210672
5.33e+08 3.304456
5.50e+08 3.387133
5.67e+08 3.514676
5.83e+08 3.634609
6.00e+08 3.636984
6.17e+08 3.453297
6.33e+08 3.09098
6.50e+08 2.620621
6.67e+08 2.051623
6.83e+08 1.398647
7.00e+08 0.6288801
7.17e+08 -0.2089145
7.33e+08 -0.8729058
7.50e+08 -0.8983268
7.67e+08 -0.1494044
7.83e+08 0.8073187
8.00e+08 1.548733
8.17e+08 1.943862
8.33e+08 2.050232
8.50e+08 1.890077
8.67e+08 1.512847
8.83e+08 0.9992631
9.00e+08 0.4794029
9.17e+08 0.180576
9.33e+08 0.1580454
9.50e+08 0.4193046
9.67e+08 0.7895336
9.83e+08 1.223261
1.00e+09 1.625476
1.02e+09 2.006417
1.03e+09 2.338812
1.05e+09 2.613677
1.07e+09 2.839776
... 61 more lines ...
|
|
|
220
|
Thu May 25 15:36:54 2023 |
Ryan Debolt, Byran Reynolds | preliminary error tests (PUEO) | Here are some preliminary results from testing the effect of error on growth in the GA. For this test, we start with a simulated error of 0.5 because our true fitness score is bounded between 0.0 and 1.0. From here, we simulate doubling the number of neutrinos by reducing the error by root(2), then root(4), and observed the growth on the fitness scores plots. We did these three tests with both 50 and 100 individuals (plots starting with 30 use 50 and starting with 60 use 100). Looking at the plots, we can see that the only plot that shows growth is 100 individuals with halved error (corresponding to 4 times the number of neutrinos). In fact, it took going to root(25) (corresponding to 25 times the number of neutrinos) to observe comparable growth with 50 individuals. This leads us to believe we likely need to increase both neutrinos and individuals to see meaningful growth in our GA. Further tests are required to see how consistent these results are. Another test, though unrealistic, we ran 1000 individuals with the root(25), this test was optimized in around 10 generations, this could be seen as a maximum performance standard. |
| Attachment 1: 30_10_10_0_48_25_6_2_half_error_fitness.png
| 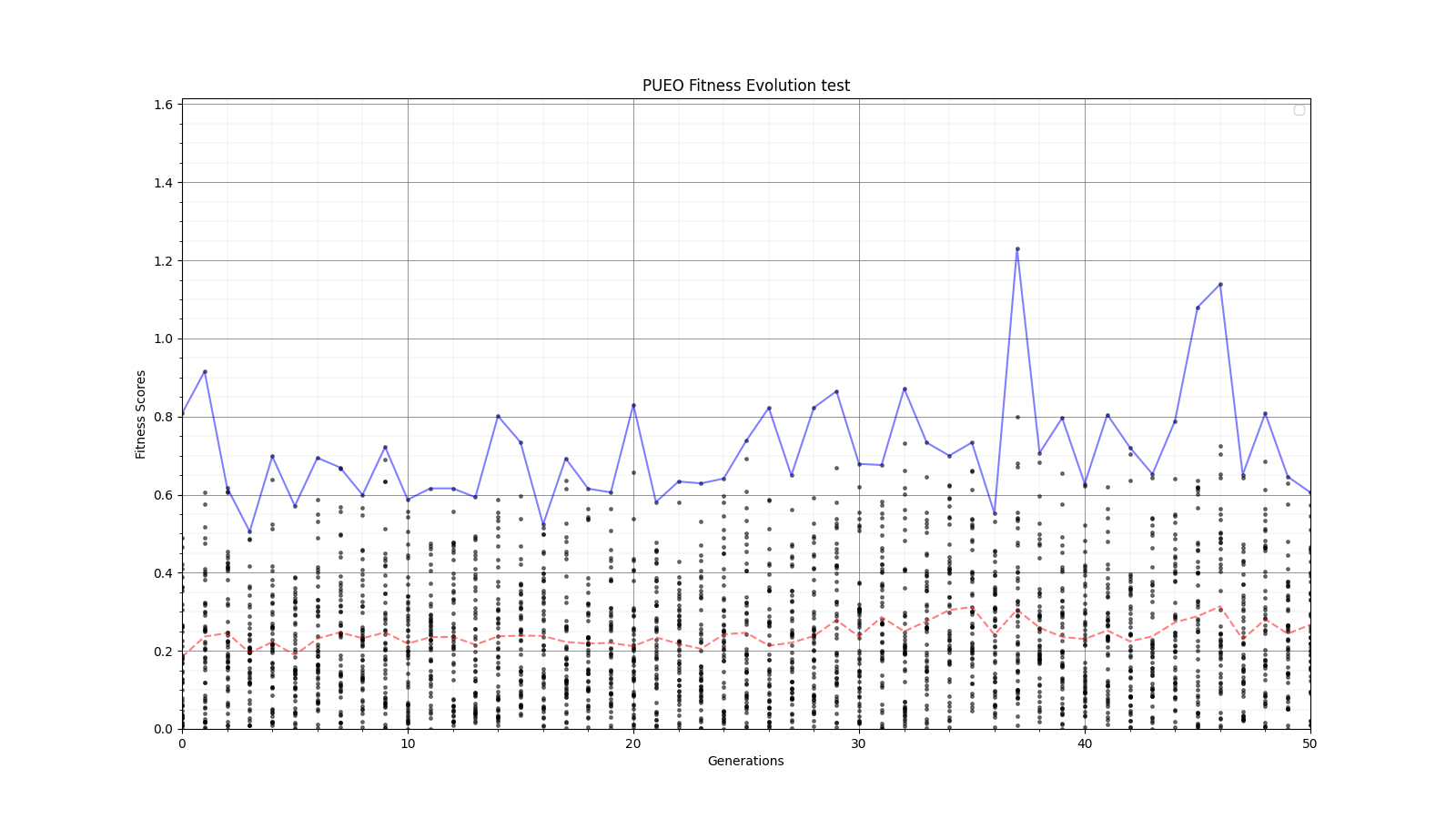 |
| Attachment 2: 30_10_10_0_48_25_6_2_high_error_fitness.png
| 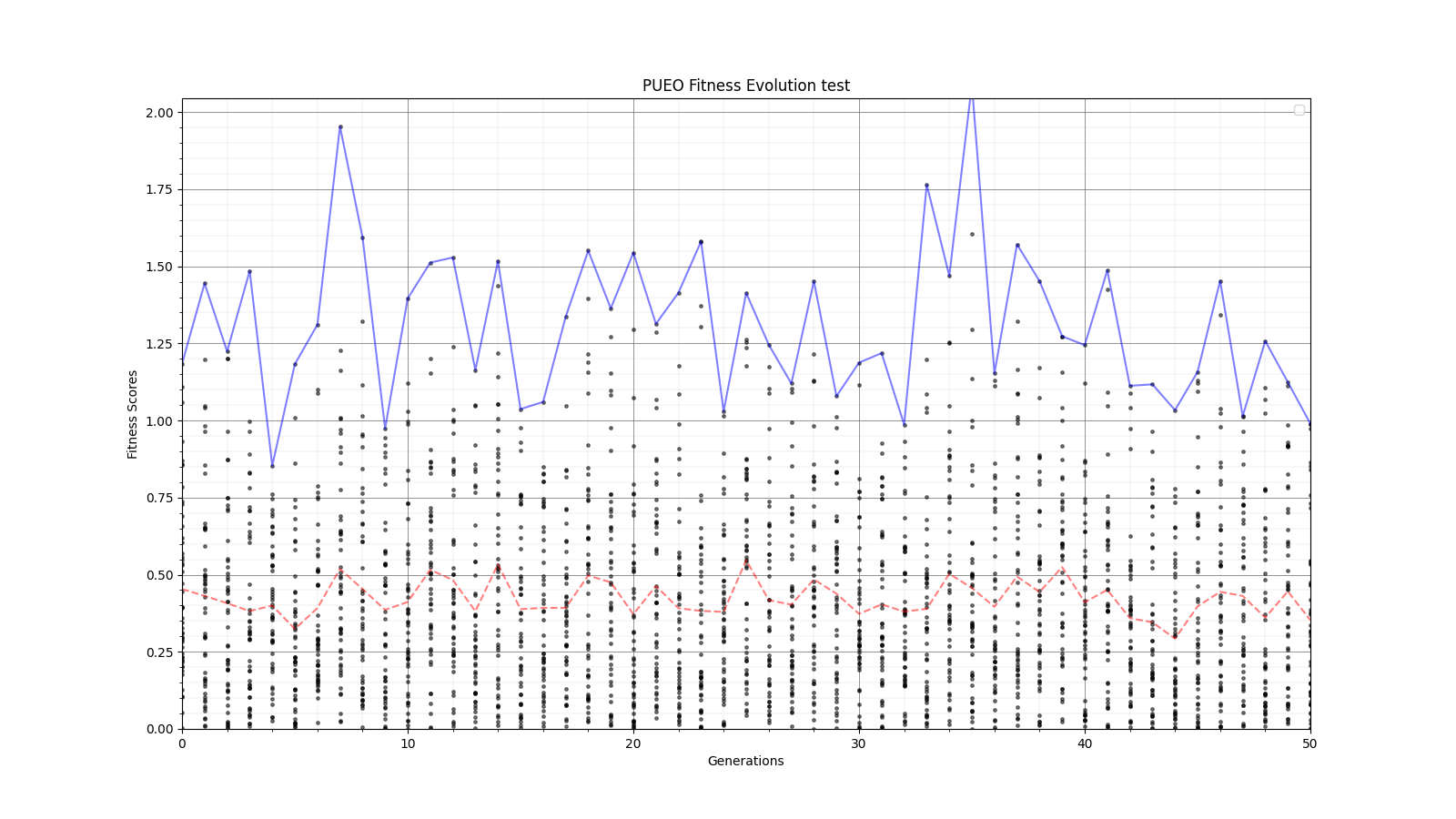 |
| Attachment 3: 30_10_10_0_48_25_6_2_root2_error_fitness.png
| 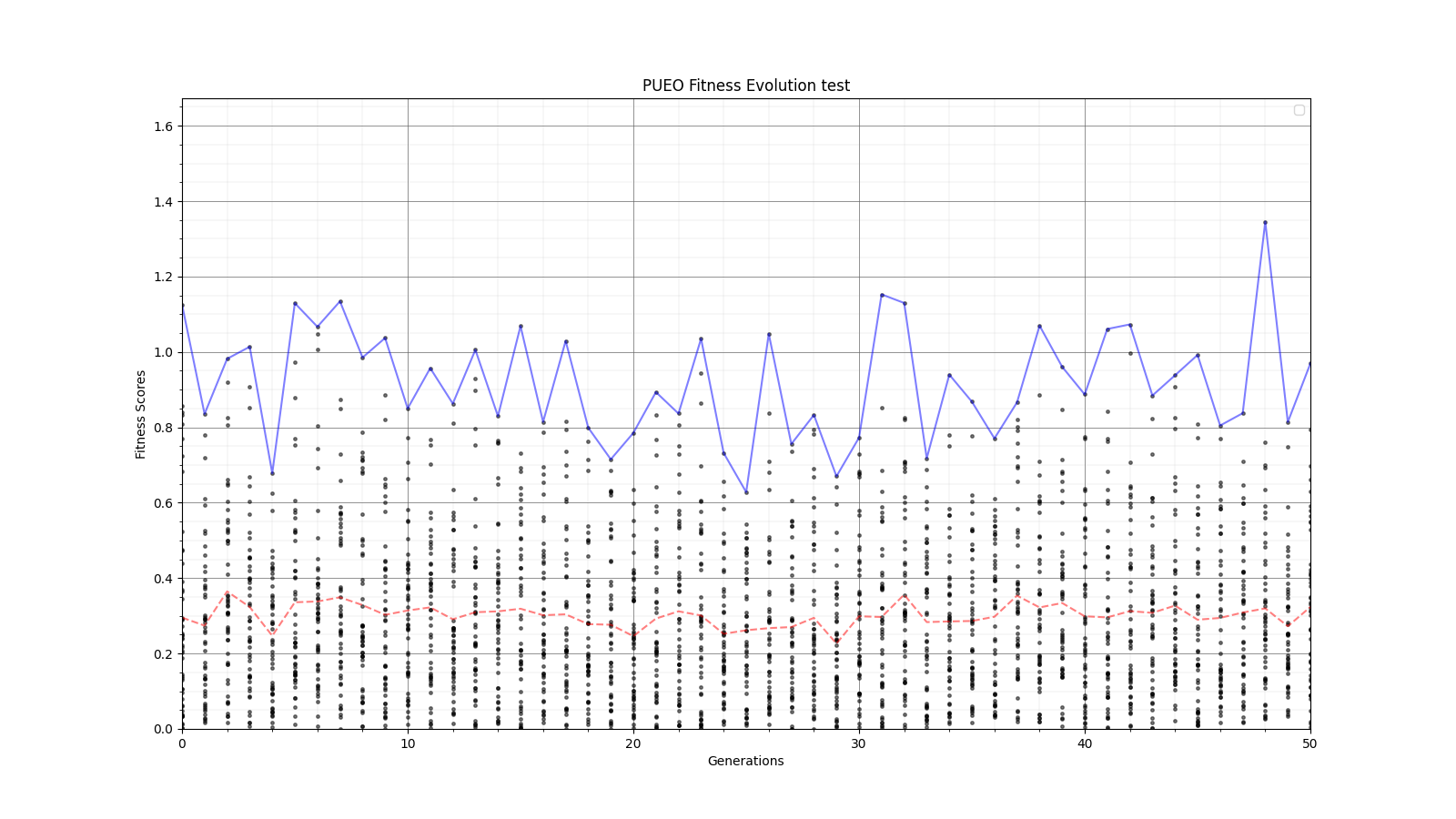 |
| Attachment 4: 30_10_10_0_48_25_6_2_root8_error_fitness.png
| 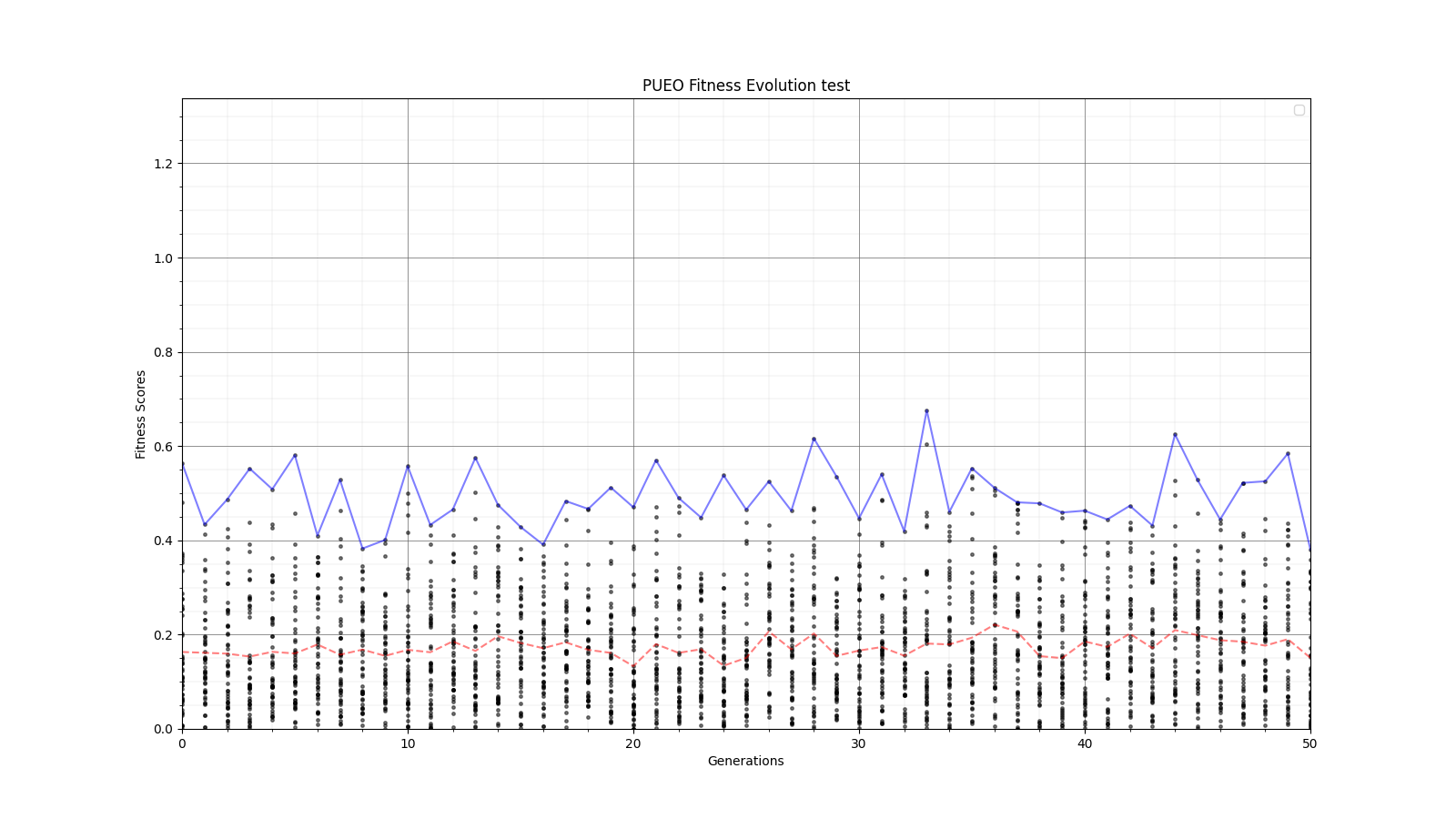 |
| Attachment 5: 30_10_10_0_48_25_6_2_root25_error_fitness.png
| 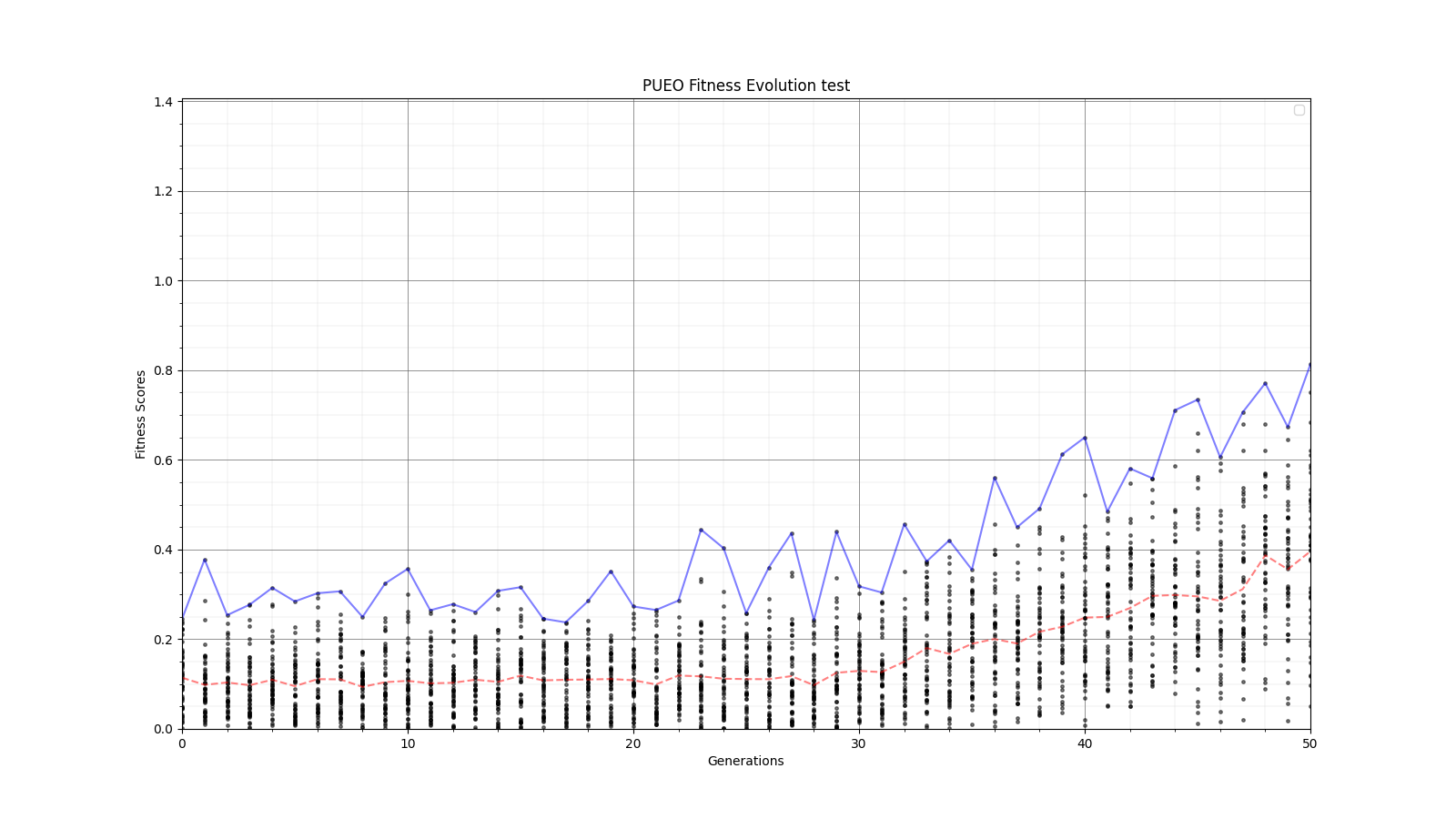 |
| Attachment 6: 60_20_20_0_96_25_6_1_half_error_fitness.png
| 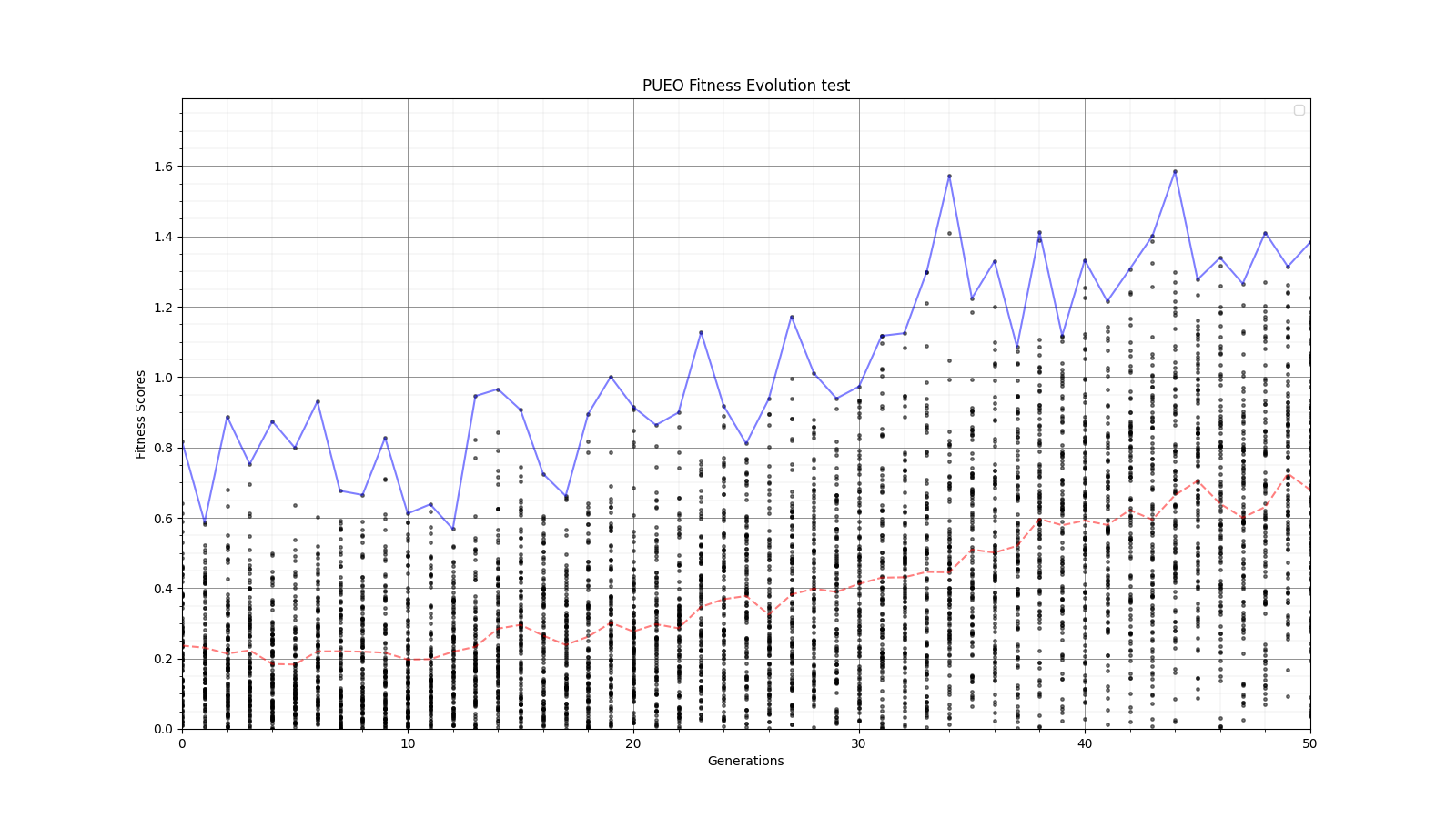 |
| Attachment 7: 60_20_20_0_96_25_6_1_high_error_fitness.png
| 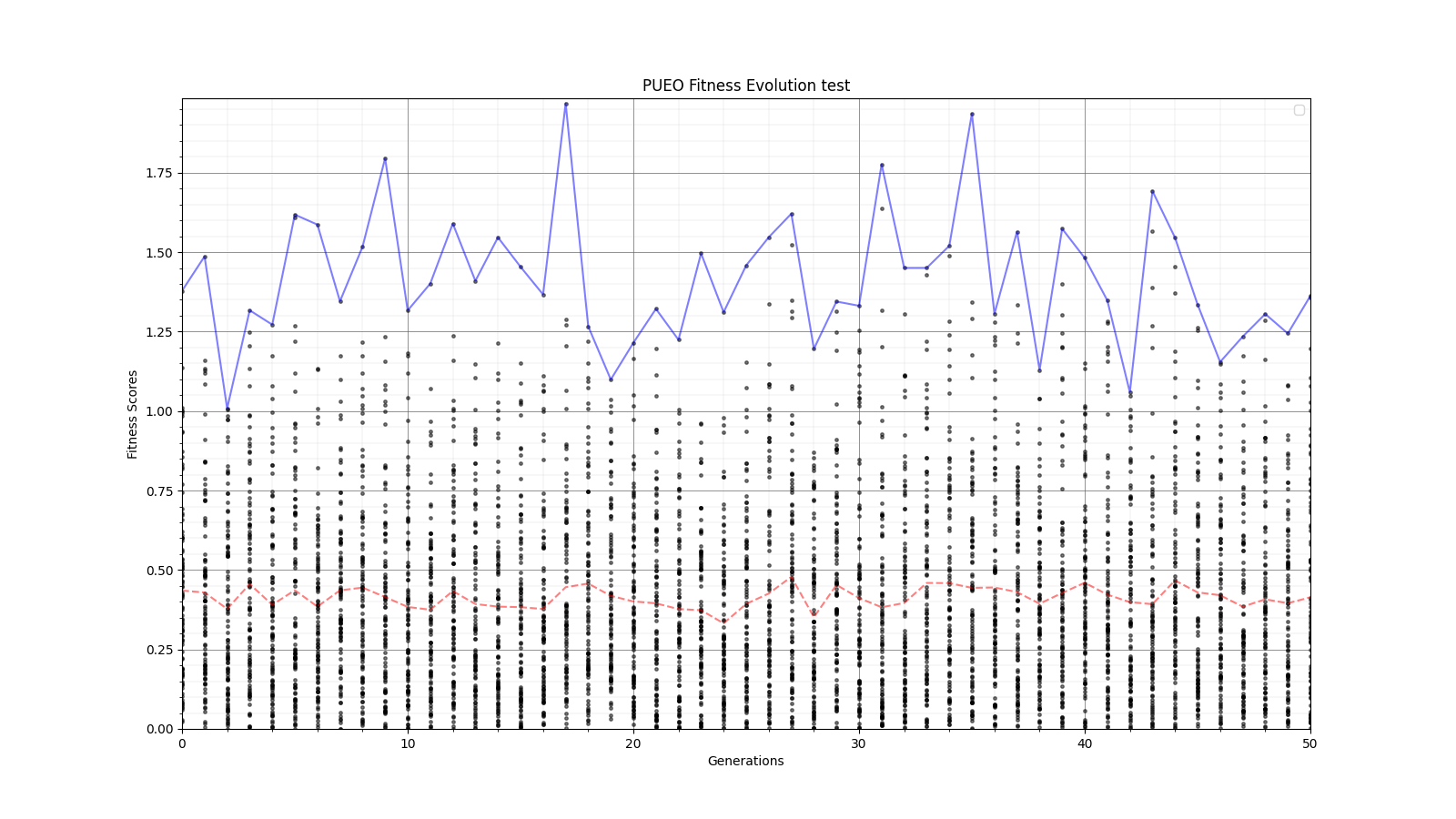 |
| Attachment 8: 60_20_20_0_96_25_6_1_root2_error_fitness.png
| 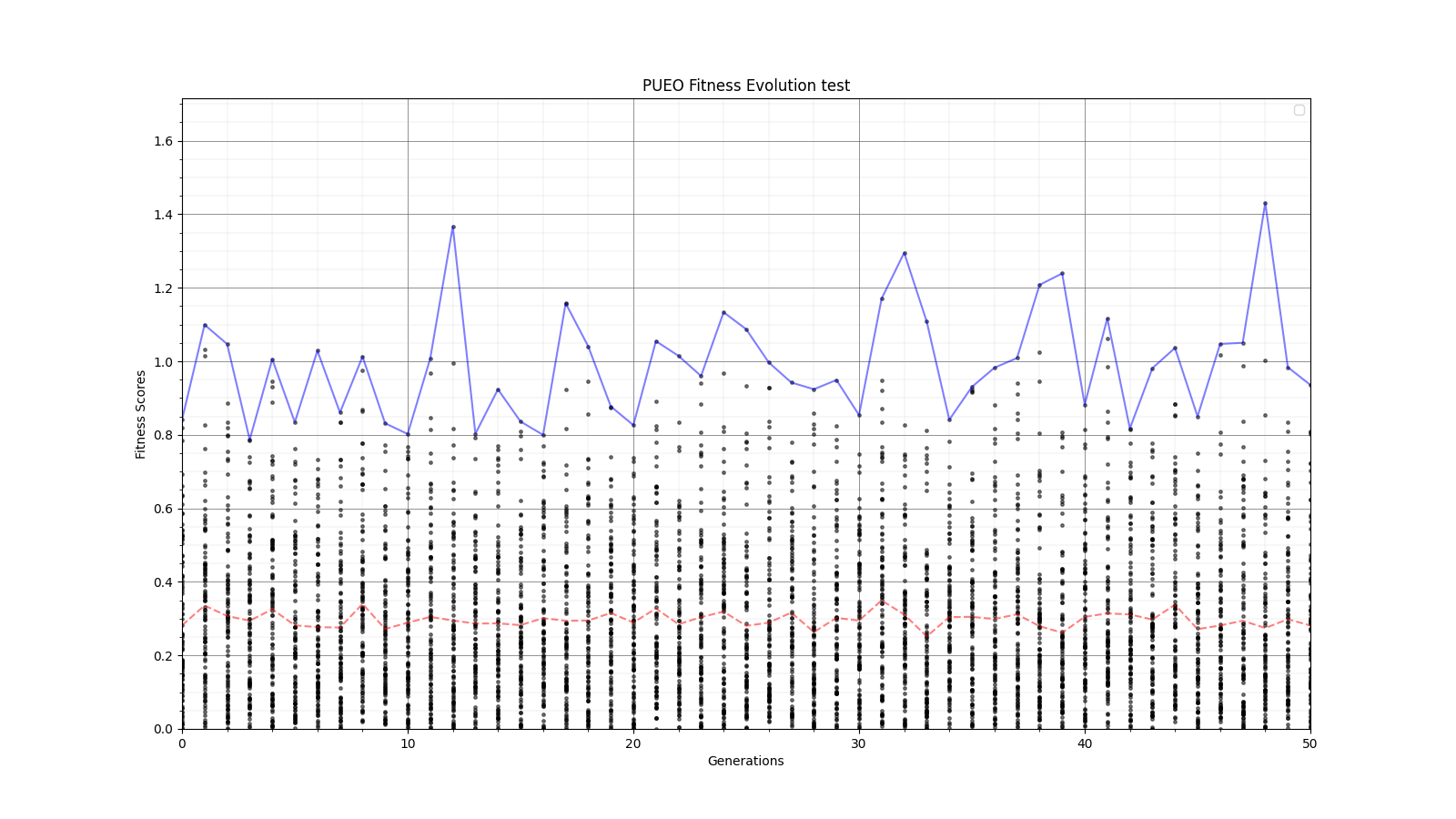 |
|
|
203
|
Mon Mar 20 13:35:25 2023 |
Amy | info on format for PUEO antenna files | William Luszczak 1:30 PM
This is the directory with the current PUEO antenna data files: https://github.com/PUEOCollaboration/pueoSim/tree/main/data/antennas. Each type of antenna will need several files:
- vv_0 and hh_0 describe the on axis v and h pol gain as a function of frequency. Gain is listed as a multiplicative factor (not in dB, if I'm remembering correctly)
- hv_0 and vh_0 describe the on axis cross polarization gain
- vv_az and hh_az describe the off-axis gain via multiplicative factors of the boresight gain. These are currently listed as a function of frequency for reference angles of 0,10,20,30,40,50,60,70,80, and 90 degrees, though we can always adjust the reference angles if needed.
- vv_el and hh_el are similar to above, but for gain as a function of elevation angle instead of azimuthal angle.
So for example, the total vpol gain in a particular frequency bin for a signal incident at azimuthal angle az and elevation angle el would be vv_0(f)*vv_az(f, az)*vv_el(f, el)
New
1:31
We can also potentially adjust this if it would be more convenient for the GENETIS people to output antenna information in a different format (or at different reference angles or whatever). The important thing is that the boresight h, v, and cross pol gains are all defined, as well as the off-axis response (as a function of azimuth and elevation) |
|
|
231
|
Wed Jul 12 21:30:56 2023 |
Alex Machtay | Zoom training on XF 07/12/23 | Recording of training on XFdtd by Alex Machtay on July 12, 2023.
https://osu.zoom.us/rec/share/FVKiTcQrRh8NLNYJADwugy0H2VjDo8ZlBsH16sJAGv2JINNwxxzyNfo5ueqVgOTO._XErF57Kz8KOM3a5
This link will expire in 120 days so note that we need to post a permanent recording.
Here's the GitHub repository with examples of Xmacros: https://github.com/Machtay/XFdtd_Scripts |
|
|
149
|
Fri Mar 25 17:32:25 2022 |
Alex M | XFdtd Step Size Investigation | The reviewer for the paper we recently submitted mentioned that our step sizes at which we are measuring the gain in XFdtd may be too large. They pointed out that there appear to be "lobes" at 400 MHz. I ran the antenna we presented in the paper through XFdtd using a step size of 5 degrees (what we've been doing) and a step size of 1 degree (the reveiwer's recommendation). Attached are antenna responses for these two different step sizes at 200 MHz and 400 MHz. Qualitatively, there are noticeable differences in the "jaggedness" of the 400 MHz plot and how extreme the minima and maxima appear, but the basic shape is preserved. We can try to do a more quantitative analysis (ex: run these through AraSim), but doing so may be more time intensive than necessary considering that AraSim may require 5 degree steps and that adding these images to an appendix may be sufficient anyway. |
| Attachment 1: polar_plot_400.06_step_5.png
| 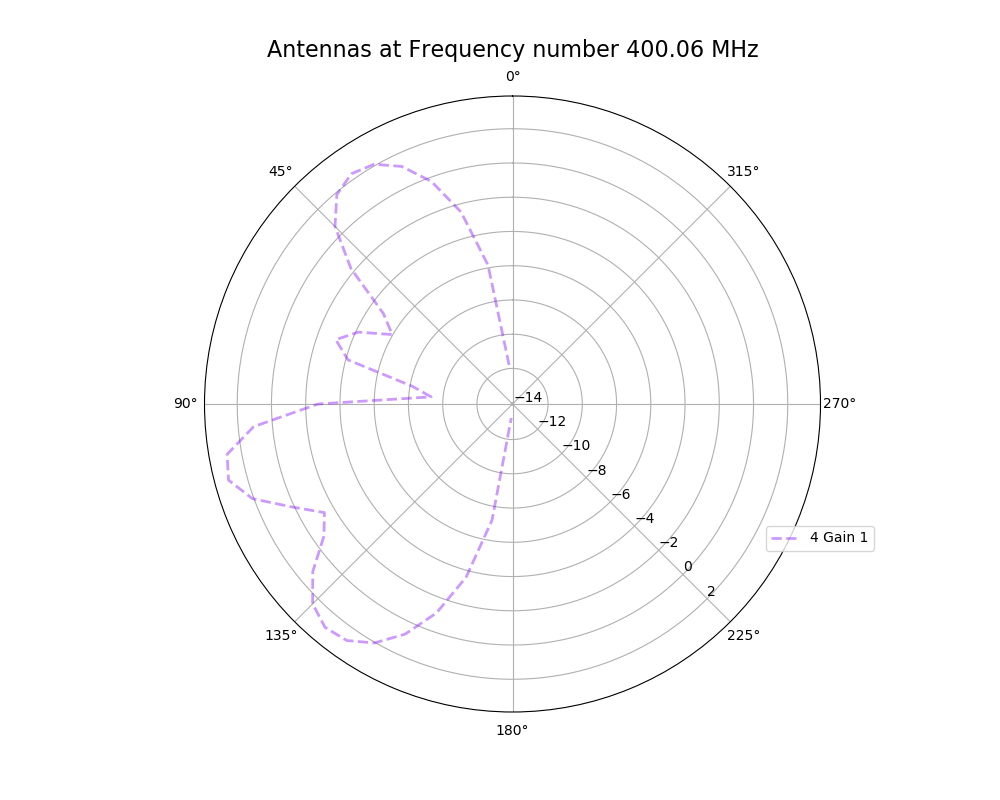 |
| Attachment 2: polar_plot_400.06_step_1.png
| 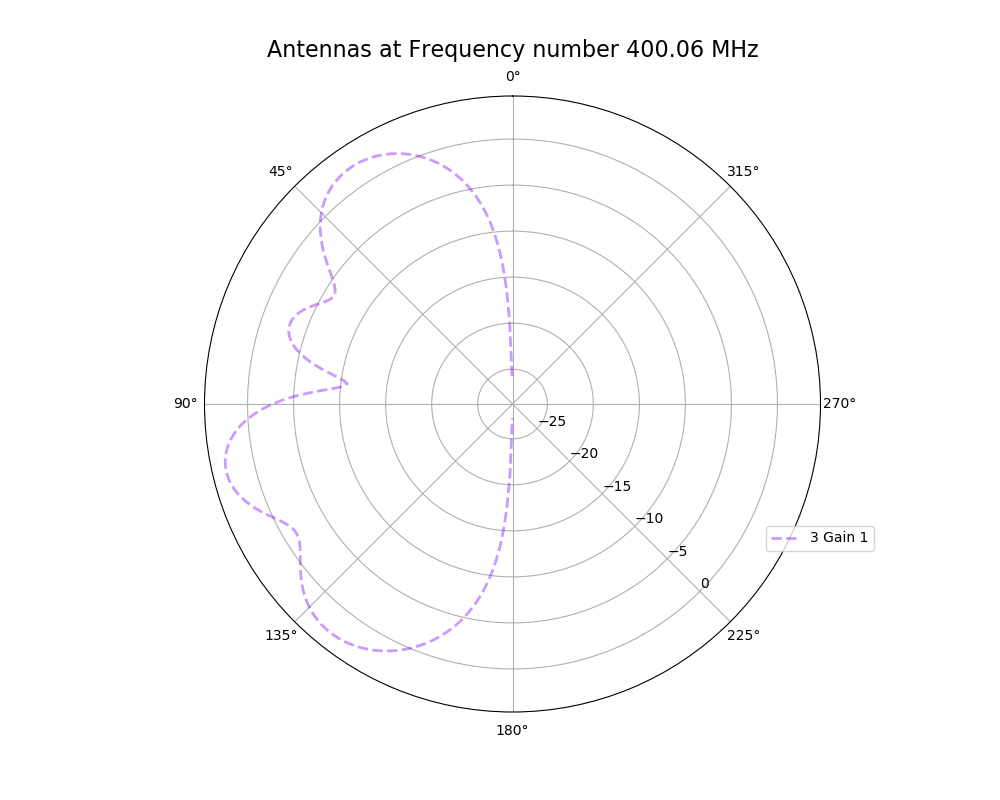 |
| Attachment 3: polar_plot_200.02_step_5.png
| 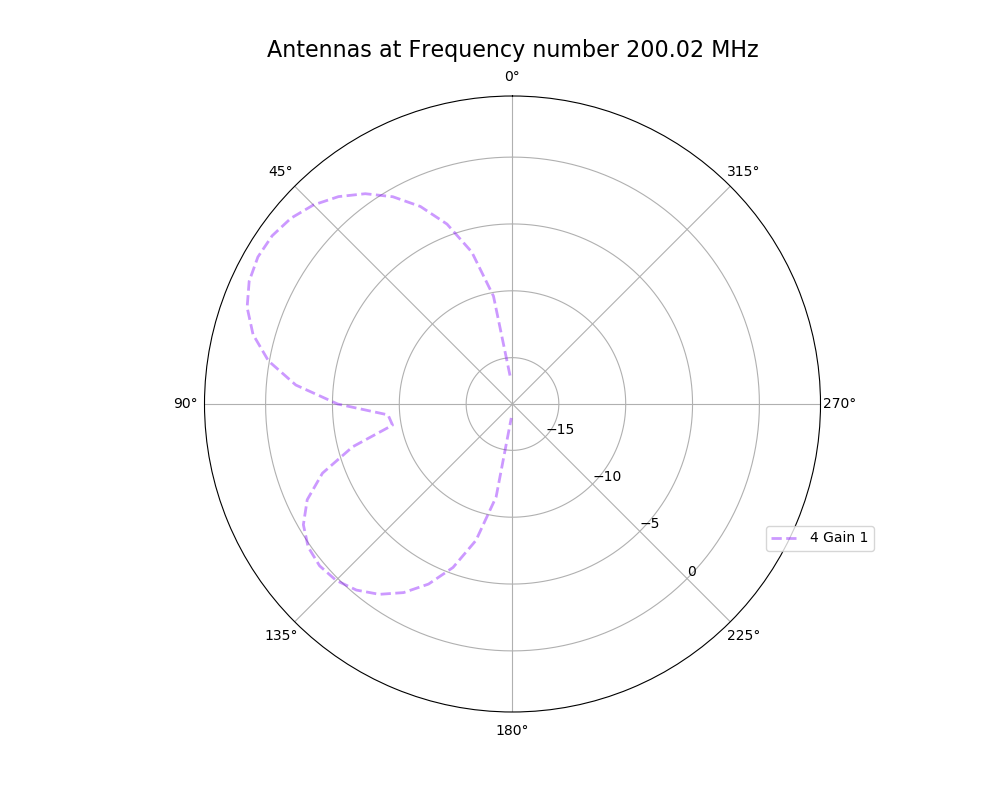 |
| Attachment 4: polar_plot_200.02_step_1.png
| 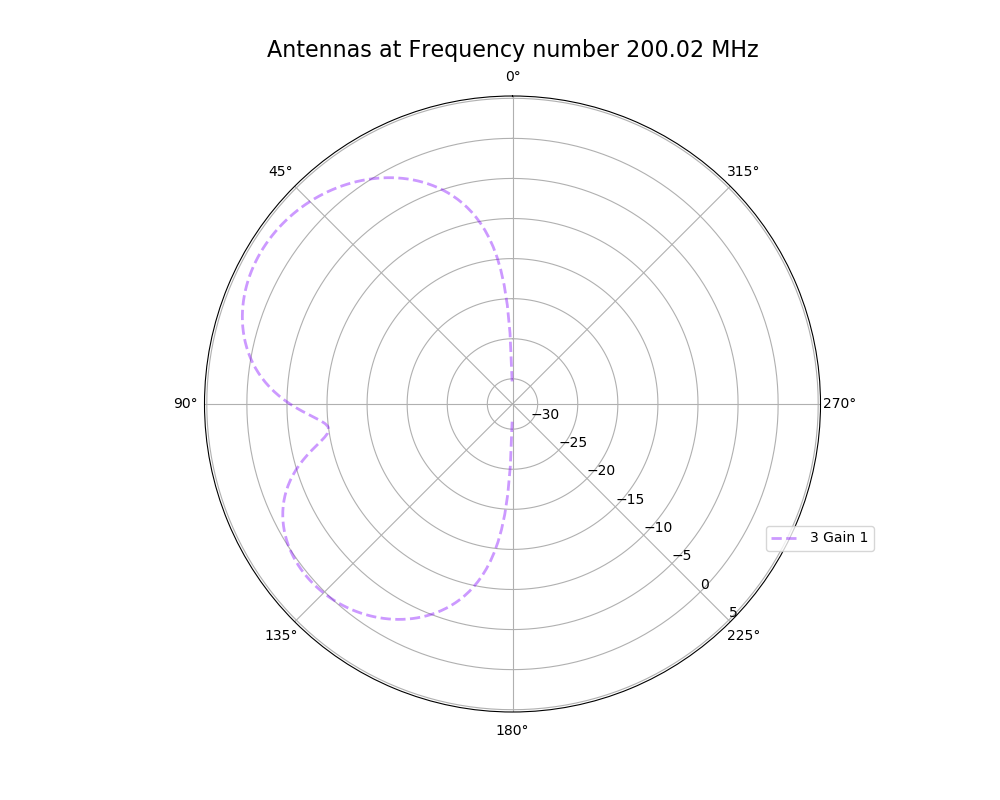 |
|
|
12
|
Fri May 17 15:13:50 2019 |
Julie Rolla | XF key flash drive holders | We have 2 physical XF flash drive keys in existence. These are non-replaceable and cannot be lost! Until further notice, the keys are held by:
(1) Cade Sbrocco (sbrocco.6@buckeyemail.osu.edu )
and
(2) Abasi Brown (brown.7146@buckeyemail.osu.edu) |
|
|
162
|
Wed Jun 8 14:45:52 2022 |
Alan S | XF Simulations | ARA bicone in ice | PowerPoint Slides | Beam patterns with ARA bicone contained in a cylinder of air surrounded by a shell of ice.
|
| Attachment 1: XF_ARA_bicone_in_ice_and_air_column.pptx
|
|
|
235
|
Wed Aug 2 00:17:32 2023 |
Jacob Weiler | XF Antenna Drawing Progress 08/02/2023 | I've been building in XF and have ran into some issues but currently have the following completed:
- Struts are placed, though they are not evenly spaced and the same. I have contacted XF to try and figure out how to get rotation/patterns to do what we need
- Circuit components and circuit board are in the xf file, though the components are not on the circuit board (I'm not totally sure how to put them on, that's the next step). The circuit components should be of spec, I looked up the part numbers from the previous ELOG post and grabbed the values from the manufacturer.
Here are the things that have not been completed/need answered:
- How to evenly space the struts connecting the shape (as stated previously, I've contacted XF to figure out how to do this easily. Im just bad at modeling in XF)
- Add circuit components to circuit board
- Add voltage and ground connections to the circuit board
- On the circuit diagram, figure out J1 and J2 connections. Meaning, which one connects to the signal from the cable and which connects to the other cone that is used as ground?
To see current antenna build go to: " /users/PAS1977/jacobweiler/GENETIS/XFzone/straightened_antenna_for_Jack.xf "
Attached are pictures in case there are issues pulling up model in XF. :)
|
| Attachment 1: sidepic.PNG
|  |
| Attachment 2: circuitboardplacement.PNG
|  |
| Attachment 3: fullpic.PNG
|  |
| Attachment 4: struts.PNG
|  |
|
|
Draft
|
Tue Mar 24 15:38:50 2020 |
Julie Rolla | X11 Forwarding Issue for Windows | Most of our students
(1)Ubuntu doesn't come with X11 forwarding on its own. So you can install Xlaunch
xeyes -display :0
|
|
|
26
|
Tue Dec 3 15:26:42 2019 |
Julie Rolla | Working session 12/3/19 | Attendance: Julie, Alex M., Alex P., Cade, Scott.
What we have done today:
(1) Fixed Concerns regarding copying the output .uan files properly so they are not overwritten. (see #8 in this elog entry: http://radiorm.physics.ohio-state.edu/elog/GENETIS/22 for details on the error). This was corrected in part E as:
for i in `seq 1 $NPOP`
do
for freq in `seq 1 60`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${i}_${freq}.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}_${freq).uan
done
done
And in the "looping part" -- ie part G we have corrected it to say the following. Note that every time we run an iteration/generation in the loop, it writes the name of the output files the exact same (individual_frequencystep.uan). It will overwrite these files in the next generation unless we move them and rename them like so. We don't want this, because we will likely want to plot the gain patterns of the individuals we like (gain pattern data is in the .uan files).
for i in `seq 1 $NPOP`
do
for freq in `seq 1 60`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${i}_${freq}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}_${freq}.uan
done
done
(2) Added scale factor "A" (actually called "Scale Factor" in our bash script and fitnessFunction_ARA.cpp), and the generationDNA.csv as an input file (this file is where the radii of all individuals in the current generation live).
- We have added this as a variable in the fitnessFunction_ARA.cpp. It now runs as a flag pass in when it runs in the bash script. In section E:
./fitnessFunction.exe $NPOP $ScaleFactor $AntennaRadii/generationDNA.csv $InputFiles #Here's where we add the flags for the generation
and in the looping part (part G):
./fitnessFunction.exe $NPOP $ScaleFactor $AntennaRadii/generationDNA.csv $InputFiles #Here's where we add the flags for the generation
and at the top where variables are declared:
RunName='Julie_12_2_2' ## Replace when needed
TotalGens=6 ## number of generations (after initial) to run through
NPOP=4 ## number of individuals per generation; please keep this value below 99
FREQ=60 ## frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=10000 ##Number of Neutrinos Thrown in AraSim
ScaleFactor=1.0 ##ScaleFactor used when punishing fitness scores of antennae larger than holes used in fitnessFunctoin_ARA.cpp
Note that every time the roulette algorithm runs, it's written into generationDNA.csv. We copy those right before rerunning the roulette alg in the next generation -- ie cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv -- so that each generations parameters don't get overwritten. However, the current generation's parameters live in generationDNA.csv until the roulette alg runs in the next generation, so we can read in that file to the fitnessFunction_ARA.cpp file to determine the constraint (ie Rantenna)
(4) Implemented Alex P.'s version of his optimized AraSim. This is now fully functional in the loop.
NOTE: WE HAVE MADE A MISTAKE! WE MADE THE CONSTRAINT THE R-VALUE WE READ IN FROM THE GENERATIONDNA.CSV FILE. THIS IS NOT THE RADIUS WE THINK IT IS! WE NEED TO DO SOME TRIG ON IT TO FIX IT!!
|
|
|
25
|
Tue Dec 3 01:26:52 2019 |
Julie Rolla | Working session 12/2/19 | Attendance: Julie, Alex P., Cade, Alex M., Mitchell, Evelyn (Go team!)
I. Adding Constraints on the in-ice hole size.
Today we worked on adding constraints on the drilled hole size. Our plan is to do so by penalizing the fitness scores of indiviuals with a radius larger than that of the hole in the ice. Ie in fitnessFunction_ara.cpp we would have:
if Rantenna>=R hole
fitnessScore =fitnessScore x (e-[A(Rantenna-Rhole)/Rhole)]^2)
else
fitnessScore= veff (or whatever way we decide to calculate the fitness score in the future, if this ever changed)
Where "A" is a constant we can adjust to tweak how heavy the penalty is. Other things to do in order to make this work:
(1) Make constant "A" a variable in the bash script.
- Do this the same way $NPOP is used in the roulette algorithm cpp program! :)
(2)Read in the roulette algorithm output file for your radius of the antenna.
- Note that this isn't a super obvious thing. Right now our roulette alg outputs are saved as $(generation)_filename.csv. So, for the first gen -- ie gen 0-- it will be 0_filename.csv. For the second gen -- ie gen 1-- it will be 1_filename.csv...etc. This means we need to make sure the fitness score calculator has a way of knowing what generation we are on, in order to be sure it is grabbing the correct radius for each individual (not that of an old generation).
We will be working on this more in our meeting tomorrow. As for now, the game plan has been to make comments on what we should be adding in the correct sections of fitnessFunction_ara.cpp (and will be doing the same in the bash script shortly, too --ie XF_Loop.sh).
II. Adding Ara Values to Plot.
We haven't started this yet, but it is something we need to do. When we plot Veff vs Generation, Theta vs Generation, and R vs Generation we need to add in the value of these for the current ARA bicone as a baseline for our evolutions.
Note: All of the contents over the last month need to be added to the manual! DO NOT FORGET! |
|
|
Draft
|
Tue Feb 25 17:10:36 2020 |
Alex Machtay & Alex Patton | Working Meeting 2/25/20 |
Today's Tasks
Mitchell, Evelyn, Eliot, Ryan: Work through the bash project; Mitchell is done, Ryan and Evelyn are close to being finished.
Leo: Worked on next presentation (not sure when he'll present--either Friday or next Tuesday).
Alex M: Continued commenting code and helped Alex P. with changes in Part_B and Roulette_algorithm .
Alex P: Fixed errors in loop, tested with an interactive job (using CPU). Errors look to be fixed up through part B. Arasim should be launching and using the $tmpdir, though we need to get further in the testing to see for sure.
Made a copy of the loop that functions without GPUs so that if GPU is unavailable some progress can be made even if it is slow.
While waiting for a GPU Alex P ran on CPUs, at half scale factor one indiviudal in XF took 32 minutes (although it was predicted to be over 2 hours).
|
To Do List:
We want to keep testing the loop. Right now, we are using a cpu running at half scale. This gives estimated times for each antenna of up to 2 hours. GPUs are suddenly hard to come by, so if we are relegated to using CPUs we'll probably need to scale down considerably.
Evelyn, Ryan, and Mitchell are probably going to be ready for the python project soon. If someone is around on Friday to help them they should be able to finish the bash project.
One idea is to run all the inital xmacros in the interactive job and then submit a GPU job that runs all the xfsolver command. With the hope that not having the GPU in the interactive job will make it easier to get the interactive job and to get the GPU for a shorter time because it would only be needed for a smaller segment. This would especially help with running larger amounts of individuals.
|
|
|
119
|
Fri Nov 20 17:50:05 2020 |
Alex M | Working Meeting 11/20/20 |
| Alex M |
We worked with Amy to track down the error we were getting with the AraSim fix. Alex P and I had gotten AraSim to compile, but running gave the error (at the bottom). We did a binary search for the source of the error and determined it comes from the if statement around line 640 in Report.cc:
if ( event->Nu_Interaction[0].LQ > 0 && (fabs(viewangle-signal->CHANGLE_ICE)<=settings1->OFFCONE_LIMIT*RADDEG) ) { |
| Ryan |
Kept testing different rations of tournament and roulette. A trend seems to appear that while full roulette was the worst run overall, it rises until a ratio of about 8R:2T with a max score of 99.22% match and then it slowly falls from that point. Need to do more testing to verify averages and to fix strange segmentation faults in crossover when running 6T and 8T ratio. |
| |
|
| |
|
| |
|
| |
|
| |
|
AraSim EField project Error:
*** Break *** segmentation violation
===========================================================
There was a crash.
This is the entire stack trace of all threads:
===========================================================
#0 0x00002b845aeb445c in waitpid () from /lib64/libc.so.6
#1 0x00002b845ae31f52 in do_system () from /lib64/libc.so.6
#2 0x00002b84562bebf4 in TUnixSystem::StackTrace() () from /cvmfs/ara.opensciencegrid.org/trunk/centos7/root_build/lib/libCore.so
#3 0x00002b84562c13ea in TUnixSystem::DispatchSignals(ESignals) () from /cvmfs/ara.opensciencegrid.org/trunk/centos7/root_build/lib/libCore.so
#4 <signal handler called>
#5 0x000000000044fd70 in void std::vector<double, std::allocator<double> >::emplace_back<double>(double&&) ()
#6 0x00000000004c3c0e in Report::Connect_Interaction_Detector(Event*, Detector*, RaySolver*, Signal*, IceModel*, Settings*, Trigger*, int) ()
#7 0x0000000000436eb3 in main ()
===========================================================
The lines below might hint at the cause of the crash.
You may get help by asking at the ROOT forum http://root.cern.ch/forum
Only if you are really convinced it is a bug in ROOT then please submit a
report at http://root.cern.ch/bugs Please post the ENTIRE stack trace
from above as an attachment in addition to anything else
that might help us fixing this issue.
===========================================================
#5 0x000000000044fd70 in void std::vector<double, std::allocator<double> >::emplace_back<double>(double&&) ()
#6 0x00000000004c3c0e in Report::Connect_Interaction_Detector(Event*, Detector*, RaySolver*, Signal*, IceModel*, Settings*, Trigger*, int) ()
#7 0x0000000000436eb3 in main ()
===========================================================
|
|
|
29
|
Thu Jan 9 14:24:25 2020 |
Julie Rolla | Winter Break Updates and new to-do list (with deadlines) | The following items from our to-do list have been successfully completed. Below this list, you can find the remaining urgent items for our proposal due date of 1/28 (items must be completed with plenty of time to spare).
Completed items:
*Note* please check the highlighted item. This one still must be verified.
- Made software plot Theta VS Gen
- Fixed current L and R plots
- These now have correct axis labels, a legend, and connect data for each individual between generations.
- Produced Veff for ARA's actual bicone parameters.
- Made XF_Loop.sh submit a job for the ARA gain AraSim input file in BiconeEvolutionOSC/AraSim/Ara_Bicone6in_outputs.txt
- STILL MUST: Make sure that the "wait function" has been properly edited to consider the submission of this extra job in the first iteration of the loop. (Not sure if $NPOP+1 instead of $NPOP fixed it correctly) ----> CADE SHOULD LOOK AT THIS :)
- Added plotting software for Veff VS Generation (includes ARA's actual bicone Veff as generated in #3).
- Fixed the ARA hole constraint in fitnessFunction_ARA.cpp -- ie fixed the calculation to consider the correct radius "r". See http://radiorm.physics.ohio-state.edu/elog/GENETIS/28 for details.
- Successfully made energy a variable in setup.txt.
- used the same "sed" format as before in the bash script (XF_Loop.sh).
- Pushed to Github.
Items to still do:
- Save CAD models in XF: Due 1/15 ------>(Cade-- second most urgent one)
Stop plots from popping up during run: Due 1/15 ------>(Alex P.)- Add "save state" idea to loop: Due 1/15 ------> (Cade and maybe Alex P can help -- This is the most urgent one)
- Figure out why fitnessFunction_ARA.cpp isn't plotting: Due 1/15 ------> (Julie, and Alex M)
- Run the loop and evolve for results: Due 1/24 ------>(Alex M, Mitchell, Cade)
- Write proposal: Final Due 1/28 ------> (Amy, Kai, Julie)
When submitting plots, please make a new folder in there and label it "Date_of_run, Energy, Generations, Individuals". For example, my folder within this dropbox link would be: "1_10, 10^19, 10, 5". I have made sample folders in this dropbox link if needed.
You can export the plots from OSC to your machine (using CP command) or can type "display NameOfPlot.png" then screenshot and crop your plot to upload to dropbox. |
|
|
20
|
Wed Nov 20 00:40:49 2019 |
Julie Rolla | Week 11/19/19 Update | Attendance for today's session: Cade, Julie, Alex M, Mitchell, Scott.
Tasks/to-do list:
- Get loop to run up to the fitness function.
- Right now we have the bash script setup to run AraSim by submitting them as parallel jobs. Today Cade added the "wait" function within the job in order to make sure the loop doesn't continue to the next step before the Arasim data is ready. Cade will give us an update on Thursday letting us know whether or not this completed or gave an error.
- Fix error at fitness function.
- Currently, the loop stops at the fitness function calculation, and aborts (program fitnessFunction_ARA.cpp). We are unsure of why at the moment. Here are the things we are looking into.
- Run this outside of the loop-- ie compile the fitnessFunction_ARA.cpp (g++ -std=c++11 fitnessFunction_ARA.cpp -o fitnessFunction.exe), and run the executable(./fitnessFunction.exe). What happens?
- Julie and Alex did this. It gave a segmentation fault. We need to look into this further.
- We double-checked that this .exe was properly created from compiling, and it was.
- Double-check our AraSim output by checking:
- Is there an error past the fitness function, and it's killing the loop after this runs?
- Is his output not properly going into the gen alg?
- Are the output files properly:
- Filled
- Formatted
- Put in the right place
- Look at how it's calculating the fitness score.
Since it seems like Alex and I may have found a problem at our first "test", this could be the issue. We will investigate it further. If (1) Cade's "wait" function works, and (2) we can fix this issue with the loop stopping at the fitness score calculation, ideally this means that the loop is then running.
Today we also found that it is better to run on the terminal through an interactive job than a virtual desktop. This is MUCH faster -- ie we don't have to wait nearly as long as we do to get a node via a virtual desktop. This means that everyone with a PC needs to either (1) work out their issues with X11 forwarding crashing (Mitchell succeeded at this today by making sure Xquartz was set up to work via ssh to Pitzer in the settings), or (2) dual boot Linux. It seems that Mac users are not having issues, but that some of the Windows users are having their OS completely crash when XF GUI tries to forward. Users can test whether or not their X11 forwarding is working by doing the following:
- Logging onto Pitzer
- Typing "module load xfdtd"
- Then typing "xfdtd"
This should successfully open the GUI for XF. If it crashes, users may have to do one of the things I mentioned above. Once we do succeed at having this setup, you can start an interactive job via the command:
qsub -I -X -l walltime=1:00:00 -l nodes=1:ppn=10
You then have to wait until the node is free. This has taken about 1/3 of the time to get than when requesting a virtual desktop. Note that, however, we may need a higher ppn (maybe something like 40). This is not totally clear yet. |
|
|
19
|
Fri Nov 15 09:59:27 2019 |
Julie Rolla | Week 11/15/19 Update | Attendance for today's session: Cade, Julie, Alex M, Mitchell, Scott.
Tasks worked on:
- Continue to fix continuity of loop: Cade and Julie worked on fixing the following errors:
- In section D of the loop, it currently
- Moves the output from the python conversion to $AraSimExec directory.
- Renames it as a .txt file (note the $i is renaming each in line with $NPOP).
for i in `seq 1 $NPOP`
do
mv evol_antenna_model_$i.dat $AraSimExec
done
for i in `seq 1 $NPOP`
do
cp evol_antenna_model_$i.dat a_$i.txt
done
However, this wasn't working and it was only properly renaming individual 1. To check this,
- We confirmed it is properly iterating the first loop by using the "echo" command. The second loop was not properly iterating, however.
- We were able to fix it by combining these steps. This was confirmed to have worked.
for i in `seq 1 $NPOP`
do
mv evol_antenna_model_$i.dat $AraSimExec/a_$i.txt
done
2. We got this iteration $i to work in the job script -- ie as an input file variable. The loop now successfully submits AraSim as a job with the proper input files.
- Clean bash script
- Mitchell and Alex M worked again today on turning every function possible into functions (and putting them in their own .sh scripts that are being called by XF_Loop.sh).
- The loop runs the first generation "manually" -- ie each step was spelled out, and nothing is put into a loop. The second+ generations then run as a loop. This is because we need to initialize the first XF run -- which then has no problems in the later iterations.
- Alex M. and Mitchell are now turning each section A-F into individual functions and put into their own .sh scripts. Thus section A would just be calling another .sh script. In gen 2+ we then loop over these SectionA.sh through SectionF.sh functions. All of these just got completed. He just needs to add the change Julie and Cade made today, and clean some things up.
- For example, he has an "if else" statement that tells it whether or not it's the first (XF initializes) generation, or in generation 2+. Cade has a few changes he would like Alex to make to clean up this part of the run.
- Idea: The loop used to run the first generation "manually" -- ie each step was spelled out, and nothing was put into a loop. The second+ generations then ran as a loop. Since we were able to get XF to import the xmacro scripts automatically -- as well as save the project with the correct name (matching the expected name initialize in the bash script)-- they've gotten rid of that first "manual" iteration, and are now fully running all generations as a loop. We haven't tested whether or not we can make this change yet, but it is worth looking into now that some changes have been successfully made with XF.
Quick note from Julie:
I tried to run from the terminal once Cade left. I get the following output/error that we should make sure isn't happening for other users (since most of this is a permissions issue that I thought we fixed. I can look into the other errors and see if anyone else gets them). For whatever reason, Cade is not having the same output errors; he is able to run the same copy just fine.
Roulette algorithm initialized.
./XF_Loop.sh: line 136: output.xmacro: Permission denied
./XF_Loop.sh: line 137: output.xmacro: Permission denied
./XF_Loop.sh: line 138: output.xmacro: Permission denied
./XF_Loop.sh: line 139: output.xmacro: Permission denied
./XF_Loop.sh: line 140: output.xmacro: Permission denied
./XF_Loop.sh: line 146: simulation_PEC.xmacro: Permission denied
./XF_Loop.sh: line 147: simulation_PEC.xmacro: Permission denied
./XF_Loop.sh: line 149: simulation_PEC.xmacro: Permission denied
./XF_Loop.sh: line 158: simulation_PEC.xmacro: Permission denied
Opening XF user interface...
*** Please remember to save the project with the same name as RunName! ***
1. Import and run simulation_PEC.xmacro
2. Import and run output.xmacro
3. Close XF
Lmod has detected the following error: The following module(s) are unknown: "xfdtd"
Please check the spelling or version number. Also try "module spider ..."
It is also possible your cache file is out-of-date; it may help to try:
$ module --ignore-cache load "xfdtd"
Also make sure that all modulefiles written in TCL start with the string #%Module
./XF_Loop.sh: line 172: xfdtd: command not found
./XF_Loop.sh: line 177: cd: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie/Julie.xf/Simulations/000001/Run0001/: No such file or directory
./XF_Loop.sh: line 178: xfsolver: command not found
./XF_Loop.sh: line 177: cd: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie/Julie.xf/Simulations/000002/Run0001/: No such file or directory
./XF_Loop.sh: line 178: xfsolver: command not found
./XF_Loop.sh: line 183: xfdtd: command not found
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/AraSimTestJob.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric/AraOut_%num%.txt’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/XF_Loop3.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_C.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_A.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957221’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957221’: Operation not permitted
Resuming...
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/AraSimTestJob.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric/AraOut_%num%.txt’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/XF_Loop3.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_C.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_A.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957221’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957221’: Operation not permitted
Resuming...
NOTE: You may want to change the following in your job for Owens
nodes=1:ppn=1 defaulting to mem=4571MB
Please review the Owens documentation:
https://www.osc.edu/supercomputing/computing/owens
Feel free to contact oschelp@osc.edu if you have any questions.
8485416.owens-batch.ten.osc.edu
rm: cannot remove ‘outputs/*.root’: No such file or directory
NOTE: You may want to change the following in your job for Owens
nodes=1:ppn=1 defaulting to mem=4571MB
Please review the Owens documentation:
https://www.osc.edu/supercomputing/computing/owens
Feel free to contact oschelp@osc.edu if you have any questions.
8485417.owens-batch.ten.osc.edu
rm: cannot remove ‘outputs/*.root’: No such file or directory
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/AraSimTestJob.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric/AraOut_%num%.txt’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/XF_Loop3.sh’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_C.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Functions/#Part_A.sh#’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957220’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957234’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957233’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957224’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957221’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957227’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957218’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957223’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957230’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957225’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957231’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957217’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957222’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.o957226’: Operation not permitted
chmod: changing permissions of ‘/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/scriptEOFiles/AraSimCall.sh.e957221’: Operation not permitted
mv: cannot stat ‘*.root’: No such file or directory
Fitness function initialized.
0
Data successfully read. Data successfully written.
|
|
|
18
|
Tue Nov 12 15:45:33 2019 |
Julie Rolla | Week 11/12/19 Update | Attendance for today's session: Cade, Julie, Alex M, Alex P, Mitchell, Scott.
Monday was a holiday, and we will be replaceing that meeting later this week. Today we worked on the following tasks:
Tasks:
- Check continuity of loop
- Alex M will try another run tonight, and has already requested a virtual desktop.
- Install Arasim to /fs/projects/PAS0654/BiconeEvolutionOCS to be edited and messed with
- Cade is testing whether or not this is running within the loop. He had gotten a few errors, and it is not completing. It's submitting the job with the new bash script, but it is not actually running.
- Found an error. The bash script was not grabbing the correct AraSim directory. More updates to come.
- Clean bash script
- Mitchell and Alex M are putting all current functions hard-coded in into their own .sh scrips that are now being called in the main bash script.
- Update Manual
Right now Elliot and Leo have decided to focus on working with Keith. They will not be contributing to this project for the remainder of the semester. Students have been posting individual updates here: https://www.dropbox.com/home/GP_Antennas/Updates
I've also attached the meeting minutes from the call last week, in order to keep track of updates. |
| Attachment 1: GENETIS_meeting_minutes_11_|_7.pdf
|
|
|
134
|
Wed Jul 14 15:45:30 2021 |
Ethan Fahimi | Wednesday Updates (7/14/2021) |
| Ethan |
Worked with Alex on fixing a few bugs with AREA. We are trying to solve an issue where the individuals are not finishing runs (around one in every four gens with 100 individuals). We believe some individuals may be too good and are then taking more than the wall time we have given them. Alex is testing this while I am working on a script that will add all the weights in the temp_{ind}.txt files. (See weightAdder.py for more) |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
8
|
Fri Feb 22 17:09:28 2019 |
Suren Gourapura | Updates that need to be added to the manual | We are redesigning the way we simulate antennas in our loop. To do this, we changed our simulationPEC macro skeletons and our output skeleton.
To make this easier, we changed the way we name the files, from i.uan where i is the simulation number, to i_j.uan where i is the antenna and j is the frequency.
We also rewrote XFintoARA.py so it's clean. Note that before there were comments from a previous author claiming it was VERY questionably written. This has now been cleaned up. We also changed the bash script relevant to the above changes.
This change needs to be added to the manual, and until we update this update, we have not done it.
|
|
|
28
|
Thu Dec 5 16:58:55 2019 |
Julie Rolla | Updated To-Do List: | Updated To-Do List:
- Add ARA constants in plotting code: ie add Veff, theta, R for ARA. This will sit as a constant line for us to compare each evolving parameter to. For example, in the plot Veff vs Generation, we will see a constant line for ARA's Veff to see how it is evolving in time.
TASK 1: MAKE Veff PLOT. Note: One of our variables in this run is Energy. Unfortunately, we care about how Veff changes with Energy for ARAactualBicone. This means that before plotting "Veff versus Generation" we need to generate Veff for ARAactualBicone at the energy we are assigning the run.
STEP 1: Make plotting code to plot "Veff vs Generation".
- You can follow the way we make LRPlot.py.
- However, in this case, we will need to take the files /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Ouputs/$RunName/AraOut${gen}_${i}.txt -- ie the AraSim outputs for each individual in ALL generations. ie Gen 0 - ${gen} -- INCLUDING the one we created in Gen 1 for ARAactualBicone -- and plot Veff from those.
- We also want to plot error bars with Veff. The error bars for each are labeled "and Veff (water eq)" at the bottom of these AraSim output files we are already reading in for Veff.
- Note, to know how to read in Veff, you can look at how we are reading that *exact* line from the AraSim output files in on the fitnessFunction_ARA.cpp code in the Antenna_Performance_Metric directory, since we are doing it there.
- Note that since we are plotting "Veff vs Generation", we will either have to read in all the files AraOut${gen}_${i}.txt (up to how may ${gen} we are in) every time -- including the ARAactualBicone one-- or we can append them all to one .txt file. It's your choice.
- Make sure you label the units for each axis every time, and that you have a key! If not, Amy will hate the plot!
STEP2: Add Veff for ARAactualBicone as a constat in the newly made "Veff vs Gen" plotting code. Remember, as I mentioned above, we must first put the ARAactualBicone through AraSim to see Veff at the same energy as the rest of the individuals. Here's what to do:
- Edit the Bash script to submit a job that runs the gain patterns of ARAactualBicone through AraSim in the initial gen (gen 0)!
- There's a current file that exits for ARAactualBicone that has the gain patterns of the current antenna, in the correct format for AraSim to run. In the bash script (XF_Loop.sh) we need to create ONE more AraSim job submission for this antenna (as a comparison to what we see with our genetically evolved ones) just in gen 0 (the first generation).
- This current AraSim input file for ARAactualBicone exists in BiconeEvolutionOSC/AraSim/Ara_Bicone6in_output.txt
- This needs to be the input for our .job script.
- Add lines to your newly made "Veff vs Gen" plotting code that reads in the Veff output for this ARAactualBicone AraSim output file (whatever you've chosen to name it when you wrote it into the job script.
STEP 3: Move the ARAactualBicone output file into the $Run_Name directory so it is not overwritten ever.
- We will want to make sure this is not overwritten after the run ends, and we start a new one.
- We will also be using it for plotting "Veff vs Generation".
STEP 4: Correct the "wait" command for the AraSim job submissions.
- The "wait" command, on the first iteration, will have to wait for $NPOP+1 now-- not $NPOP.
TASK 2: MAKE "Theta vs Gen" Plot.
STEP 1: Make plotting code to plot "Theta vs Generation".
- You can follow the way we make LRPlot.py; however, MAKE SURE YOU LABEL AXIS WITH UNITS, AND HAVE A KEY.
- This will be exactly the same as this other code, except you will need to:
- Read in theta from your generation DNA files
- Add the ARAactualBicone constant to the code as a reference point
TASK 3: Fix "L vs Gen" and "R vs Gen" plot.
STEP 1: Correct both plots to have labeled the units for each axis every time, and a key. This is plotted in the "LRPlot.py" code.
STEP 2: Fix our 'R' values. I do believe we are plotting the incorrect "R". Here is what I think is happening:

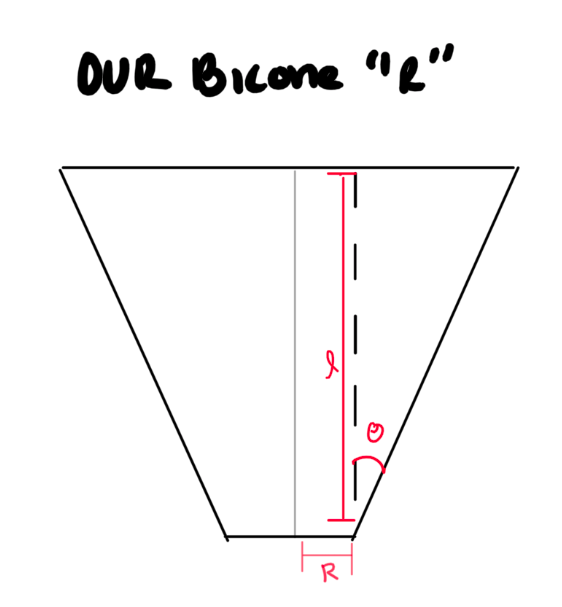
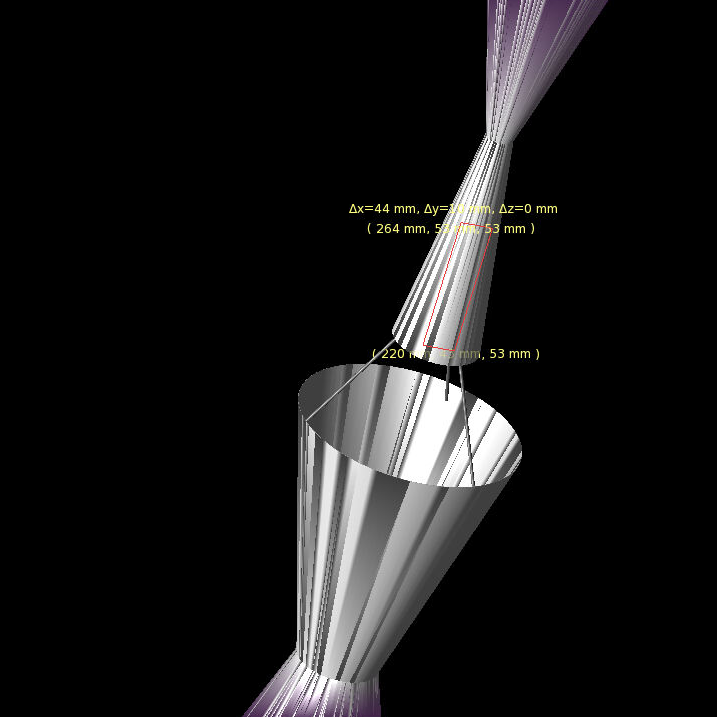
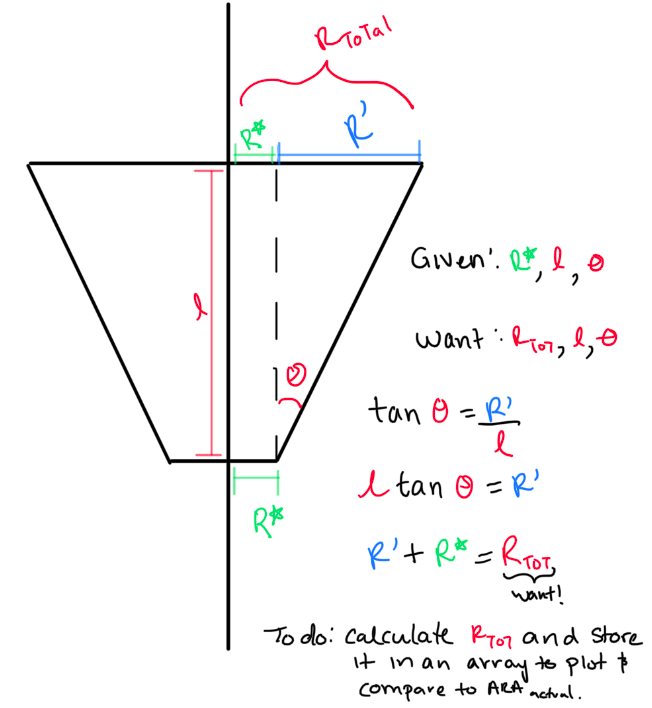
- We need to double-check and see what ARA defines "R" as on their current antennas.
- Check with Amy?
- What does Brian Clark's thesis say?
- Can you find a paper that shows a picture of how they define it?
- If we are wrong, this means we need to change the "R vs Gen" part in the "LRPlot.py" code so that we:
- Read in R, L, and Theta for each generation.
- See how this is already done for reading in R, and repeat it for L and theta.
- Do trig to calculate the same "R" as ARAactualBicone defines. See picture below!
- Store that newly calculated "R" as an array, and plot THAT.
- Don't forget to add the ARAactualBicone Radius as a constant line.
TASK 4: See PDF for other items! |
| Attachment 3: Proposal_To-Do_List.pdf
|
|
|
7
|
Fri Feb 22 15:46:33 2019 |
Julie Rolla | Updated To-Do List | We are getting to the stage where we would like to verify each step. It has been requested by Amy that we verify them visually. Here are the plots we need to make.
- Add plotting for L & R parameters (note that we already have similar software to this from the dipole check. You can find this in the GitHub from the dipole test here: https://github.com/hchasan/XF-Scripts). This should be between A, and B in the loop drawing on the manual. (Done)
- Plot the distribution of theta and phi of incoming signal. This should be after D in the loop drawing on the manual.
- Plot the distribution in cartesian coordinates of the neutrino interaction position. Maybe we can also plot the distance between the interaction point and the antenna. This should be after D in the loop drawing on the manual.
Finally, as a tasks not to do with plotting, we need to make sure we don't lose any data as we go. We want to make sure that we keep tagged or labelled which individuals survive, and save the data flagged for those as we go. |
|
|
175
|
Fri Aug 5 09:59:07 2022 |
Dennis Calderon | Updated Effective Volumes and Errors (Paper Run and Curved Sides) | Update to previous ELOG post for Effective Volumes and Errors for AraSim simulations for antennas from Paper_Run and Curved_Sides.
AraSim version compiled ~11/2021 and the (old) GENETIS version
Here are the column definitions:
- The first column indicates the antenna being described. For example, XF_model_21_20 was the 20th individual in the 21st generation for the run discussed in the paper. Curved_side_1 was the best performing individual from the curved run done in February 2022. Curved_Side_1_Quad_Zero indicates the curved individual with its quadratic term set to 0. Curved_Side_1_Straightened indicates the curved individual with the quadratic term set to zero and the linear term modified so as to keep the outer radius the same as it originally was (that is, for Curved_Side_1).
- The next three columns are the measurements for that individual.
- The first one idicates the mean effective volume of the runs done for that individual. There were ~300 runs, each of 10,000 neutrinos.
- The second column indicates the standard deviation of the mean (the previous column). This was measured by taking the standard deviation of the effective volumes (the line that says test Veff(ice) in the AraOut file) from all 300 runs (that is, Sigma(mean - x_i)/Sqrt(300)).
- example of line:
- Veff(ice) : 5.25337e+09 m3sr, 5.25337 km3sr
- The final column indicates the average of the errors on the effective volume measurements from the AraOut files (the line that Veff(water eq.)) divided by Sqrt(300) for each of the 300 jobs (that is, Sigma(x_i/Sqrt(300))/300).
- example of line:
- And Veff(water eq.) error plus : 0.552722 and error minus : 0.552722
Note: Previously, the best curved individual (according to the run data) had been excluded (instead, we had the 2nd-6th best individuals). This has been corrected, with the order corrected.
Also find attached a file called full_resimulate_DNA.csv . It contains the DNA for the top five curved individuals, the DNA for those same individuals with their quadratic term set to 0, and the DNA for those individuals with the quadratic term set to zero and the linear term modified to maintain the same outer radius as in the original run.
Finally, the three images show the original best curved bicone (Curved_Side_1), the best curved bicone when its quadratic gene was set to 0 (Curved_Side_1_Quad_Zero), and the best curved bicone when its quadratic gene was set to 0 *and* its linear gene was changed to keep the same outer radius (Curved_Side_1_Straightened).
We have chosen XF_Model_23_08 to be the one actually built for testing. This was decided based on the data presented here. XF_Model_23_08 was the highest scoring individual in the paper run. Because the paper data was generated using the old version of AraSim, it was decided that using those results would make the most sense for which antenna to build. This would also mean that we can compare directly to the best individual in the paper. This individual was chosen over the curved sided inviduals because it has a higher fitness score, both when using the old version of AraSim and the new version. |
| Attachment 1: Effective_Volumes_Errors_2.pdf
|
| Attachment 2: full_resimulate_DNA.csv
|
Hybrid of Roulette and Tournament -- Thanks to Cal Poly / Jordan Potter
Author was David Liu
Notable contributors: Julie Rolla, Hannah Hasan, and Adam Blenk
Done at The Ohio State University
Working on behalf of Dr. Amy Connolly
And the ANITA project
Revision date: 21 March 2018 1800 EST
0.08333,0.1,0.11667,0.13334,0.15001,0.16668,0.18335,0.20002,0.21669,0.23336,0.25003,0.2667,0.28337,0.30004,0.31671,0.33338,0.35005,0.36672,0.38339,0.40006,0.41673,0.4334,0.45007,0.46674,0.48341,0.50008,0.51675,0.53342,0.55009,0.56676,0.58343,0.6001,0.61677,0.63344,0.65011,0.66678,0.68345,0.70012,0.71679,0.73346,0.75013,0.7668,0.78347,0.80014,0.81681,0.83348,0.85015,0.86682,0.88349,0.90016,0.91683,0.9335,0.95017,0.96684,0.98351,1.00018,1.01685,1.03352,1.05019,1.06686
Matrices for this Generation:
6.31483,77.6017,0.00260615,-0.197494
3.97024,40.9244,-0.0103313,0.36119
2.32499,79.663,-0.00224948,0.192602
0.308809,39.6742,-0.0084247,0.40629
2.32499,75.9855,-0.00224948,0.192602
0.966617,42.4246,-0.0084247,0.40629
5.83731,79.663,-0.000594616,0.0106887
2.316,38.0641,0.00166678,0.0135261
4.66148,79.663,-0.000594616,0.00582814
1.75822,39.9608,-0.000356019,0.0167485
6.31483,77.6017,0.0,-0.197494
3.97024,40.9244,0.0,0.36119
2.32499,79.663,0.0,0.192602
0.308809,39.6742,0.0,0.40629
2.32499,75.9855,0.0,0.192602
0.966617,42.4246,0.0,0.40629
5.83731,79.663,0.0,0.0106887
2.316,38.0641,0.0,0.0135261
4.66148,79.663,0.0,0.00582814
1.75822,39.9608,0.0,0.0167485
6.31483,77.6017,0.0,0.00474767
3.97024,40.9244,0.0,-0.0616123
2.32499,79.663,0.0,0.0134017
0.308809,39.6742,0.0,0.0720468
2.32499,75.9855,0.0,0.0216741
0.966617,42.4246,0.0,0.0488755
5.83731,79.663,0.0,-0.03668019
2.316,38.0641,0.0,0.0769706
4.66148,79.663,0.0,-0.0415408
1.75822,39.9608,0.0,0.00252170
|
| Attachment 3: original_best_curved.png
|  |
| Attachment 4: best_curved_quad_zero.png
|  |
| Attachment 5: best_curved_same_radii.png
|  |
|
|
Draft
|
Mon Oct 5 17:34:32 2020 |
Leo Deer | Update Monday 10/5/2020 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
Finish up mutation function and then make sure it functions properly, currently our added functions all compile but we haven't tested if they are functioning as intended yet. Also have some questions about handling the standard deviations in mustations but first we want to get it functioning before applying more tweaks |
| Eliot |
Worked on learning about 3D plotting in python. Made a simple surface plot that can be expanded upon for our purposes. |
|
| Leo |
Fixed the averge Fscore Plot so that it connects the mean values with a line instead of making them discrete points. Then I made Kai's plot suggestion. For that I copied the current FScore plotting software and made some edits so that it takes in 2 runs and plots the fitness scores of both in either red or green along with the average fitness score of both. I'll attach images of what these 2 plots look like. |
For Friday I'm going to look over the other plotting suggestions but most likely begin on fixing the random seperation in the RLTS plots. |
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
| Attachment 1: FixedFScore.png
| 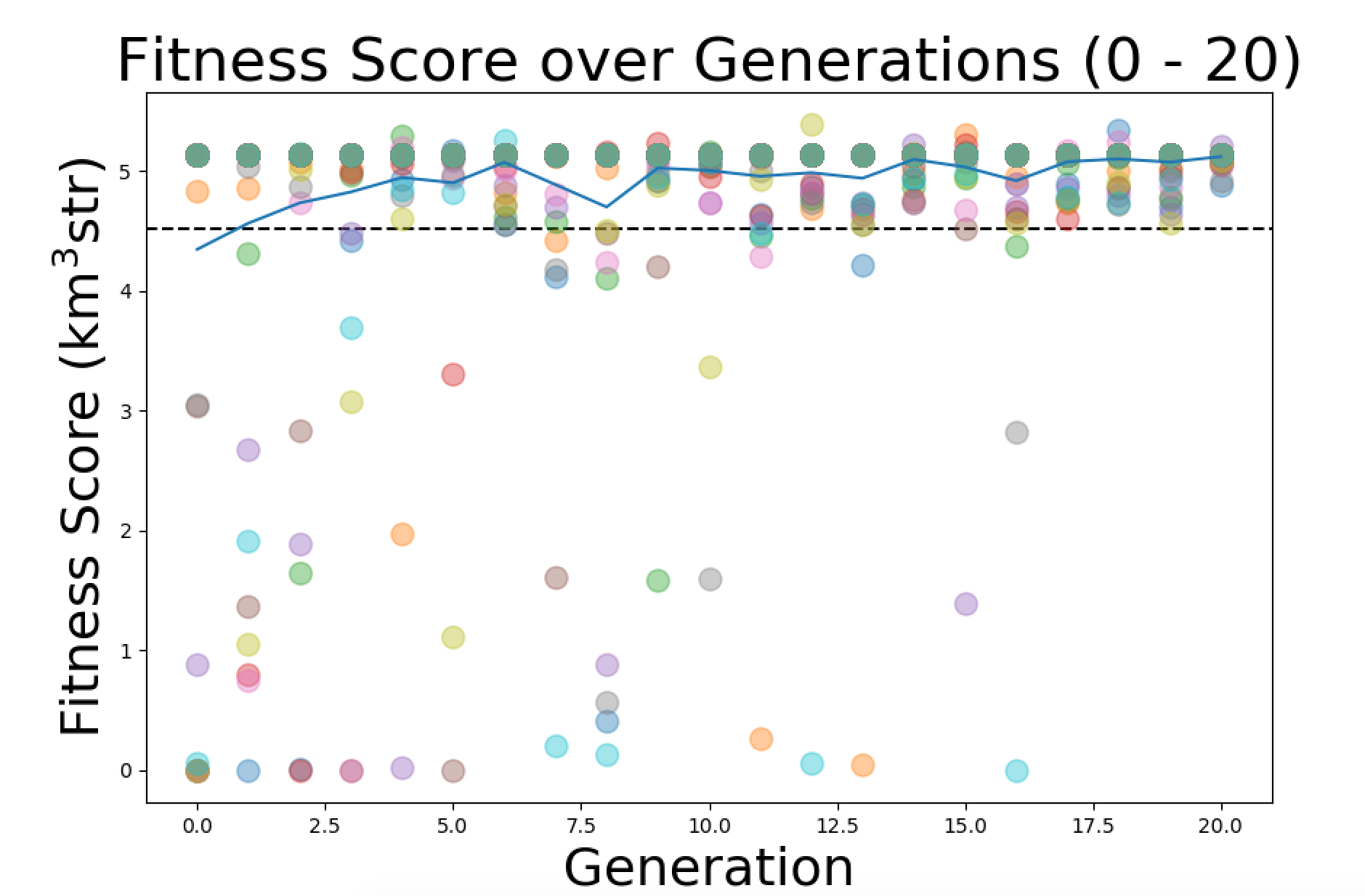 |
| Attachment 2: RedPlot.png
| 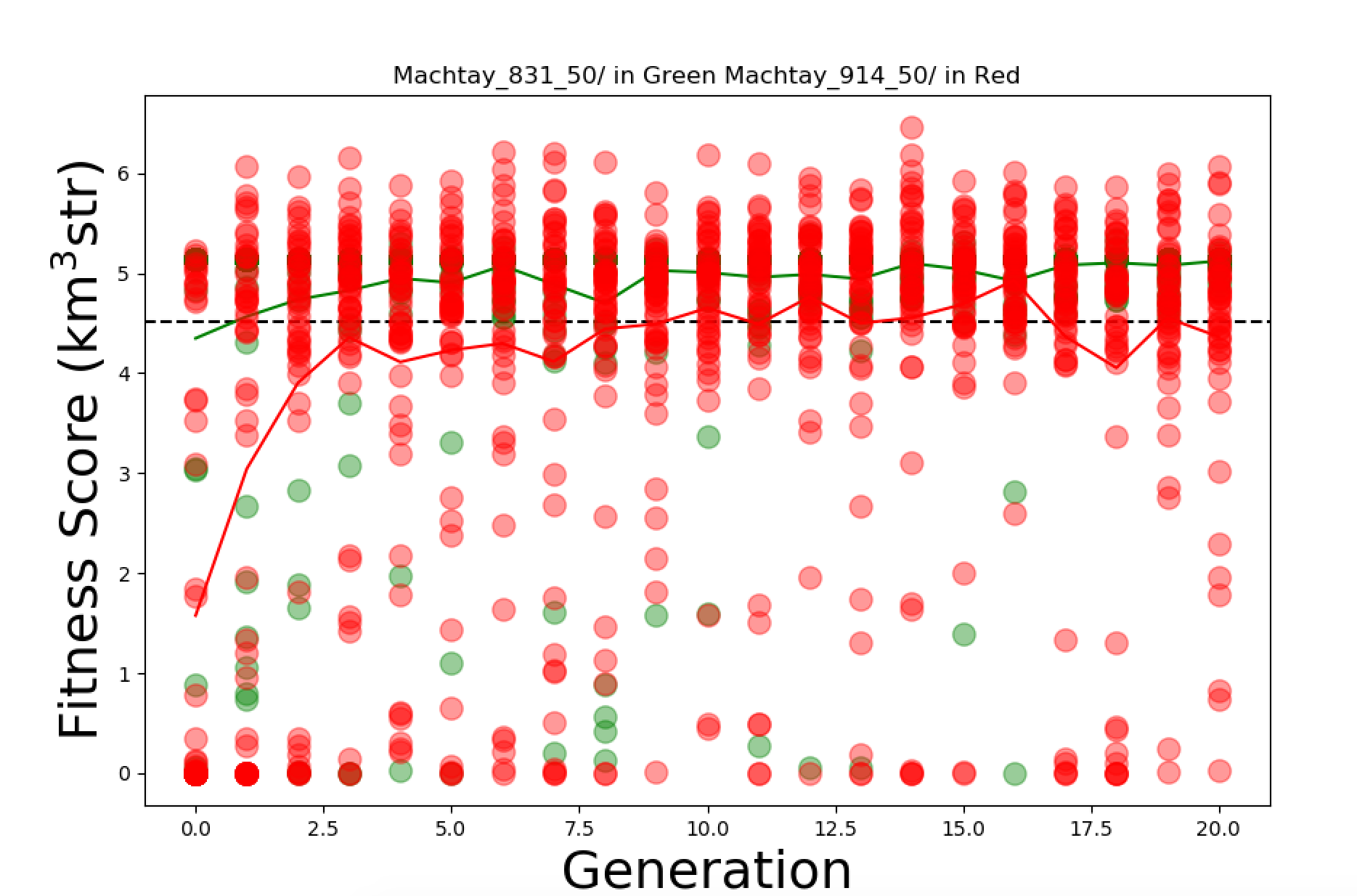 |
|
|
109
|
Fri Oct 2 17:11:25 2020 |
Ethan Fahimi | Update Friday 10/2/2020 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
Today I started working on improving the plots. I started with plotting the average fitness score for each generation on top of all the individuals data points. I made the average fitness score a star with higher contrast so it will stand out. Here is the plot of the first 26 generations of the "Machtay_20200914_Asymmetric_50_Individuals" run with the new averages. |
On monday I'm going to try and create the plot of 2 runs overlaid on top of eachother as Kai suggested. |
| Evelyn |
|
|
| Ryan |
Worked with Ben and Alex P. on making the Bicone function modular on our copy. We finished writing a new roulette selection function, tournament selection function, and reproduction function. |
Finish writing the mutation and crossover functions. After these are complete apply these functions in place of the existing function that is wrong and test to make sure the results are not producing errors. |
| Ben |
|
|
| Ethan |
Updated the AREA code with Alex M's help to work using SLURM. Began a trial run to see if our updates work. |
Begin making changes that allow the job to run faster as well as investigate what is causing some individuals to have fitness scores beyond what should be physically possible. |
|
| Attachment 1: Screen_Shot_2020-10-02_at_5.39.33_PM.png
| 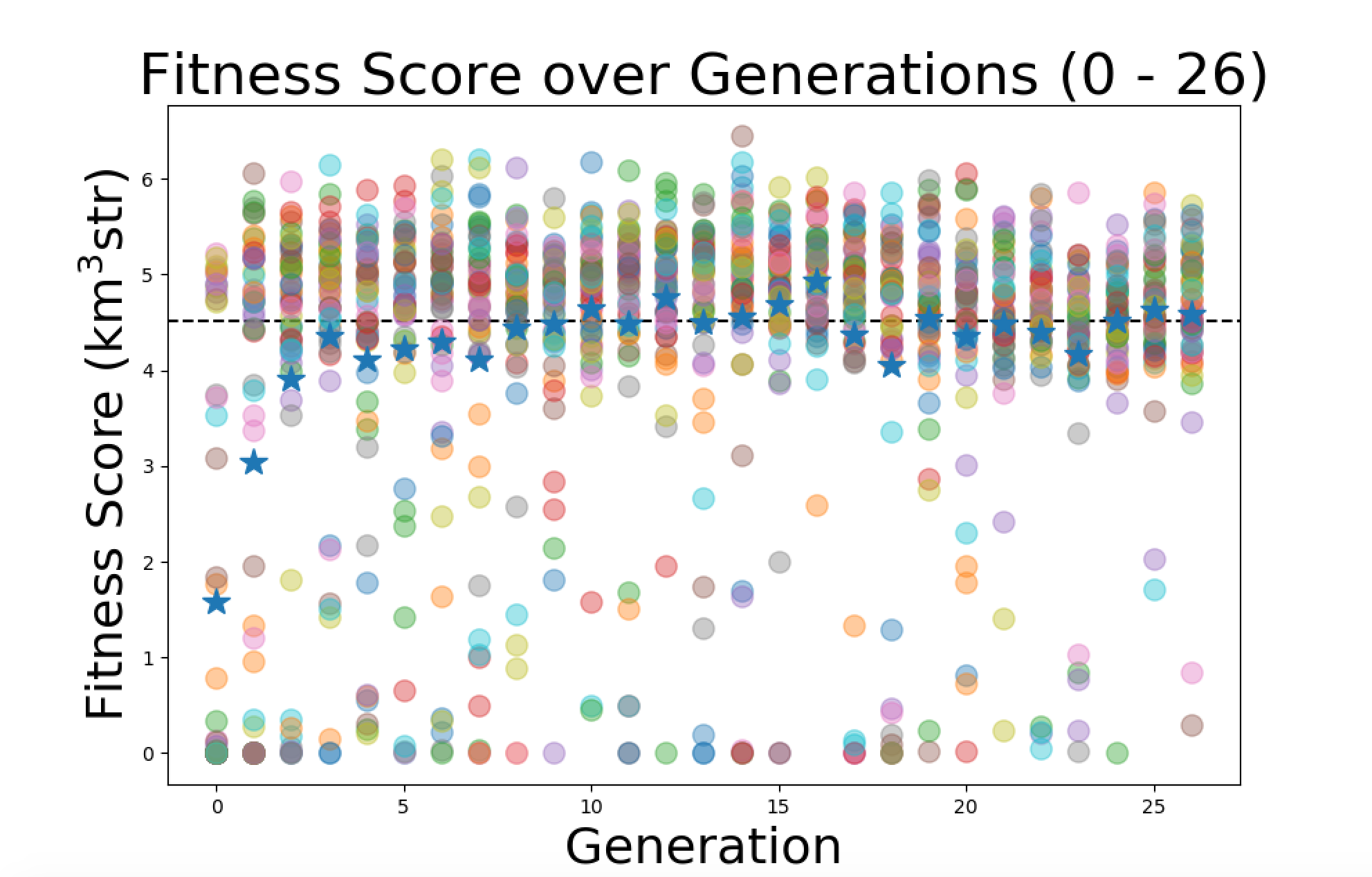 |
|
|
107
|
Wed Sep 9 17:28:54 2020 |
Alex M | Update 9/9/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I've been working on readying the loop for the transition to slurm. OSC is switching from its current job managing software (torque/moab) to slurm, which uses some different commands but is functionally similar. Since we have places where jobs are called in the script, I've been changing them (with a backup copy of those files that we know works on torque).
Right now I'm testing all of the changes I've made on the space OSC has set aside for playing with slurm. I have changes in all of the places I think they belong, but I need to make sure there aren't typos/different commands from what I've used.
|
I'll keep testing the changes I've made for slurm. I'll also try to organize some of the dfferences I've found in using slurm vs torque (for example, requesting an interactive job) so that everyone can see them and be able to find out what to use when OSC makes the switch (Pitzer changes on the 22nd, Owens is in November I think). |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
127
|
Tue Jan 26 17:17:41 2021 |
Ethan Fahimi | Tuesday Updates 01/26/2021 |
| Ethan F |
Continued implementing some solutions the OSC helpdesk gave me for fixing the unwanted extensions on the job output files. |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
219
|
Mon May 22 20:34:59 2023 |
Amy | Things to do before starting new PUEO run from 5/22/23 meeting | Here is a list of things we agreed to do before starting a new PUEO run. Feel free to add notes here as things are done and we will revisit the list next week.
Change range of the height variable to 0.75-1.75m
Start with all bad individuals so that we can be sure and see growth - all at 0.75m?
Make sure we are using the ratios in the GA that we think we are
If we are using reproduction, are we combining results when individuals are resimulated to reduce errors? I made a program to accomplish this and created a pull request on the PUEO Github (Dylan)
Investigate whether there are obvious places to speed up PUEOSim
Consolidate the GA to one branch
Understand what is happening with cross-pol, make sure we're not gettting Veff=0 when using XF-generated cross-pol response
Edit: Have the xmacros use the realized gain instead of the idealized gain, which allows for the design to build in matching
|
|
|
233
|
Mon Jul 24 15:51:28 2023 |
Ryan Debolt | Test Loop runs that need done. | |
Types of runs I need (May need some other people to help me run these):
-
Optimization runs
-
Non-error
-
Search over combinations of selection methods and genetic operators
-
Fix Sigma at 10%.
-
Do this for 10 runs for each
-
Re-run the best run for 100 tests to see if the results agree with 10 test results
-
This will be the main talking point
-
Save the plot for the best combination (proxy and metric)
-
Error
-
Re-run the best run for 100 tests with 3 different error amounts (0.1, 0.2, 0.3)
-
We will compare these results to the non-error run to talk about how errors may affect consistency/speed.
-
Save an example plot of an error test for each error (both metric and proxy score)
-
Population
-
Re-run the best run for 100 tests with 3 different population sizes (100, 500, 1000)
-
We will compare these results to the non-error run to talk about how population size affects consistency/speed.
-
Save an example plot of a population test for each size (just proxy score)
-
Demonstration runs (single runs, fitness score, no error population 100)
-
Crossover only
-
Mutation only
-
Reproduction only
-
Injection only
|
|
|
241
|
Mon Oct 7 15:35:01 2024 |
Dylan Wells | Template Run Results Slide Deck | Copy this slide deck to use when presenting on run updates!
https://docs.google.com/presentation/d/1Tk-B1QbFTP_5pQZovfn0_wbTswZTmOrJURpi374xJWo/edit?usp=sharing |
|
|
21
|
Wed Nov 20 15:05:14 2019 |
Julie Rolla | Task update 11/20/19 | Today I spent some time looking at task 1 from the update here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/20. Below are my updates.
Task-- Fix error at fitness function:
Currently, the loop stops at the fitness function calculation, and aborts (program fitnessFunction_ARA.cpp). The following are the things I have tried.
1.Run this outside of the loop: Julie and Alex did this 11/19/19. It gave a segmentation fault. UPDATE: HERE'S WHAT I DID TO CHECK THIS TODAY:
(a) Checked using cout: I used some cout commands to print sections looking for errors. It seems we were not properly giving it arguments when running it outside of the loop. Since it's running outside of the loop, these variables are not initialized. Thus, to run you need to type: ./fitnessFunction.exe $NPOP $AraOut_i.txt
- For example: ./fitnessFunction.exe 1 AraOut_1.txt
Status: The output file is verified! Running this way does properly generate our output file named fitnessScore.csv. I've double-checked this file, and it is properly being filled. At the moment it is giving a Veff of 0, thus no valid fitness score; however, this should be fixed automatically once we get real outputs from AraSim without a Veff of 0.
2. Check file location: See if fitnessScore.csv is being located in the correct place when the genetic algorithm initializes the next generation.
(a) Searched bash script for the final location of fitnessScores.cvs:
In lines 298, 299 of the bash script, it is doing the following
cp fitnessScores.csv "$WorkingDir"/Run_Outputs/$RunName/0_fitnessScores.csv
mv fitnessScores.csv "$WorkingDir"
This is taking the .csv output generated from fitnessFunction_ARA.cpp, and is moving it to the run name directory and renaming it based on the generation -- ie first gen is 0_fitnessScores.csv-- so that data isn't overwritten. Then, for some reason, it moves the original copy -- ie fitnessScores.csv-- to the location /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop
In line 374 we then run our genetic algorithm again.
./roulette_algorithm.exe cont $NPOP
This does not seem to take in the input file as an argument.
(b) Checked roulette_algorithm.cpp for the location it accesses fitnessScores.csv:
I checked if it was hard-coded into the roulette_algorithm.cpp script. I found the following lines.
// Now we need to read the fitness scores:
ifstream fitnessScores;
fitnessScores.open("fitnessScores.csv");
string fitnessRead[NPOP+2];
for(int i=0;i<(NPOP+2);i++)
{
getline(fitnessScores,fitnessRead[i]);
if(i>=2)
{
fitness[i-2] = atof(fitnessRead[i].c_str());
if (fitness[i-2]<0)
{
fitness[i-2] = 0; // If the fitness score is less than 0, we set it to 0 to not throw things off.
}
}
}
fitnessScores.close();
}
This means that our script roulette_algorithm.cpp is grabbing the fitnessScores.csv from its current directory-- which is accurate due to the line (mv fitnessScores.csv "$WorkingDir") referenced above in the bash script. Something we could verify here is to make sure this function is properly working by having it cout elements from the fitnessScores.csv within this function and to run the roulette algorithm separately. We could do this now, however, we don't have real outputs in fitnessScores.csv anyway, since we are getting a veff=0 for AraSim temporarily.
In the meantime, I've added the following line after section E (the section that runs fitnessFunction_ARA.cpp, and generates/moves/copies fitnessScore.csv):
echo 'Congrats on getting a fitness score!'
This should check to be sure that it runs until after this point. I've also added the following line after section E (section that plots the fitness score versus gen):
echo 'Congrats on getting some nice plots!'
This should check to be sure that it runs until after this point. Note that I have added these lines to both our original bash script, and the one Alex M. is editing to turn every section into a function. I have added these lines to the .sh scripts containing the functions to be sure the function scripts for these tasks ran completely. Once we have real AraSim outputs, we should be able to create some cout lines in the instructions highlighted in yellow above.
3. Look at how it's calculating the fitness score.
I have not checked this yet. However, I do not think it is likely this is the issue. I will check later though. |
|
|
13
|
Mon Jun 24 13:49:49 2019 |
Julie Rolla | Summer update | Right now the following people are working on this project:
Julie Rolla, Cade Sbrocco, Scott Janse, Alex Patton, Eliot Ferstl, Abassi Brown, Evelyn Shank, Sophie Dunlap
Today we met for our first working meeting. Scott, Alex, and Eliot were asked to speed up and optimize AraSim. Sophie and Evelyn are working on editing the .csv file with the beam patterns coming out of XF. Currently it's written in a different format than AraSim takes in. Abassi is helping Evelyn and Sophie by giving them a sample program helping reorganize the cells in a .csv file. Since there's confusion on how to optimize AraSim, we've started dissecting the software so we can understand how it works. The following are questions on the setup.txt document.
Setup.txt
What is "timestep"? Could we increase the timestep? Is this timestep big or small compared to 2 GHz?
Answer: Timestep is the unit of discrete time the computer operates at. The timestep by default is set to 2 GHz, or 5 * 10^-10 seconds. We might be able to make up some cheap time by lowering that but would have to check if it makes the data less accurate.
What is "TRIG_WINDOW"? Is this a "coincidence"? Meaning if this signal is picked up within a certain time between stations it's considered the same event?
What is "V_MIMIC_MODE"? Comments in Setup.CC says "// default : 0 - write out all chs where global triggered bin is center of the window"
Answer: Not sure exactly what it does but we can't change it from default as the other values are for the test bed mode only.
Random Question from Setup.txt, but relevant for other things. What is "testbed mode"? Answer: The "testbed" is the original test station only 40m under. This mode is for if we do detector=3 in the first part of the setup.txt file; thus, we won't use this.
What's the difference between "DETECTOR" and "DETECTOR_SIMULATION"? We found "DETECTOR_STATION" in Settings.h. The comments say "// for DETECTOR=4, indicates the single station to be simulated // 0 = testbed, 1 = A1, 2 = A2, 3 = A3"
In this file, there's an ANTENNA_MODE variable. You can look at what it does in Settings.h. In this file, it says that you can decide whether to use a bicone antenna response or "a different response for the top vpol antennas". How does this function work? What does it mean by "top vpol antennas"? Someone should look up the ARA antennas, and look into this function.
Our goal is to meet and work as a group twice a week. Collaboration in work should help us progress faster.
|
|
|
229
|
Thu Jul 6 13:12:04 2023 |
Jack Tillman | Straightened Sides AraSim Results | After discretizing and straightening the sides of the curved antenna that performed the best (Generation 13, individual 84) and running the antenna through AraSim, the following results and physics plots were generated. |
| Attachment 1: Straightened_Sides_AraSim_Results.pdf
|
| Attachment 2: Straightened_Sides_AraSim_Results.pptx
|
|
|
14
|
Mon Jul 1 13:45:49 2019 |
Alex Patton | Status on optimizing AraSim | We ran valgrind (a profiler) on AraSim to find out where it was spending most of its time, we ran it with callgrind to give an output file of the time and opened that file with qcashegrind to display it in a legible format. This let us know that the function Param_RE_Tterm_approx was taking up about 75% of the time because it was being called hundreds of millions of times. Now more specifically the standard math functions pow() and exp() were being called in this function every time and took up about 44% and 20% respectively. They were called about the same number of times as Param_RE_Tterm_approx but they are only called in the else block in an if-else statement.
JULY 31st Update:
I used the C++ library future and set up iterations of a loop to run asynchronously as they do not rely on each other's calculations. This launches a seperate thread whenever I call one. The struggle with this is that launching a thread and getting its results takes time as well, so there had to be enough calulations in a thread in order for it to make up any time. The purpose of the function GetVm_FarField_Tarray is to create arrays of doubles called Earray and Tarray and those are the only values that get altered in this function. Tarray is created with simple calculations so I left that out of threads but making Earray calls the other function that takes up so much time.
I edited the files signal.cc and signal.hh to work with 4 threads and ran 10,000 neutrinos with 4 cores. The cpu time was 1:48:41 but the real walltime it used only took 00:54:40. The time these tests for 10,000 neutrinos with base AraSim normally take anywhere from 1:35:00 to 1:45:00 or in that range. It should still work with less threads available it would just run slower and probably at the cpu time, which might be slightly slower than base AraSim but as soon as more threads are available it runs much faster as we see here. I still need to check for the accuracy of these edits, the number passed is in a reasonable range, but I will need to investigate further by giving the exact same neutrinos to both programs and making sure their outputs are identical.
I also have some further ideas to speed it up even more. Currently every iteration of the loop calls a thread but what would be faster would be to either
A: Set it up so it can call more threads at a time and thus go faster, which would be an easy edit but only work if more cores are given
B: Set it up so the current threads run more than one iteration at a time, I've started experimenting with this but having trouble making sure that everything involving pointers stays safe, but the benefit of this is that the threads would get called and joined significantly less and would cause a great speed increase without adding more threads.
All these developments are new after me spending a while trying many different ways to speed it up or run parts in parallel like this, so even if this isn't accurate or valid I believe I would be able to make it so with more time and work.
Attached I have a zip that contains the two files I edited as well as an output file from my first test. I will continue to test and improve this but am unable to work on this again until August 19th so I wanted to share my current status before I am unavailable. |
| Attachment 1: NewFilesAndResults.zip
|
|
|
129
|
Thu Mar 11 15:41:52 2021 |
Alex M | Running The Loop Updated | Because of issues we were having with XF when running at the command line, we've shifted to running through a Pitzer VDI (see here: https://ondemand.osc.edu/pun/sys/dashboard/batch_connect/sys/bc_desktop/vdi/session_contexts/new).
The VDI will give you a mirror of a desktop running on OSC's side, so things like XF will run much faster. You can open a terminal by clicking the black box at the bottom of the screen and then run the loop like normal (./Asym_XF_Loop.sh). This has the added benefit of making it possible to exit out of your browser, close your laptop, or turn off your computer without interrupting the run, since the process is running on OSC's side rather than piping to your computer. |
|
|
141
|
Fri Feb 4 16:10:32 2022 |
Julie Rolla | Run Log Template | For PAEA Algorithm:
Part I: Complete as soon as the run starts
Run details: Please answer all of the questions below!
- Run Type
- Answer here whether or not it's AREA (Gain pattern evolution) or PAEA (Bicone evolution)
- Run Date
- Run Name
- Parameters evolved
- Why are we doing this run?
- Add answer here about what we are testing
- What is different about this run from the last?
- Add answer here with info on what we are doing differently from the last run
- Example: We are testing a new operator, we are changing the percentages of each selection method used, etc.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Add answer here (say N/A if this is an AREA run)
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- Selection methods used (% of each):
- Are we using the database?
- Add answer here. This can be found in the main bash script within the variables section. (Answer N/A for AREA run)
Please upload the text file with all run details before closing this!
Part II: Complete as soon as the run ends
Results: Once this run completes, please upload the plot(s) to this post as an attachment, as well as a general explanation of the results.
- Summary and comments on results
- Add thoughts on what we should do next here!
- Example: "It seems like we are seeing minimal evolution. We should take a step back and try to see why we don't see improvement. Once we trouble shoot, we can start a new run to investigate."
- Example 2: "Run looks good! Our best individual is in generation #12. Attached are the CAD drawings of that individual."
**Upload all plots, run parameter text files (file that has run settings saved), CAD models of best indivduals, etc**
For AREA Algorithm:
Part I: Complete as soon as the run starts
Run details: Please answer all of the questions below!
- Run Type
- Answer here whether or not it's AREA (Gain pattern evolution) or PAEA (Bicone evolution)
- Run Date
- Run Name
- Parameters evolved
- Why are we doing this run?
- Add answer here about what we are testing
- What is different about this run from the last?
- Add answer here with info on what we are doing differently from the last run
- Example: We are testing a new operator, we are changing the percentages of each selection method used, etc.
- Single frequency run or run with broadband frequency dependence:
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- Selection methods used (% of each):
- Any other things to note?
Please upload the text file with all run details before closing this!
Part II: Complete as soon as the run ends
Results: Once this run completes, please upload the plot(s) to this post as an attachment, as well as a general explanation of the results.
- Summary and comments on results
- Add thoughts on what we should do next here!
- Example: "It seems like we are seeing minimal evolution. We should take a step back and try to see why we don't see improvement. Once we trouble shoot, we can start a new run to investigate."
- Example 2: "Run looks good! Our best individual is in generation #12. Attached are the CAD drawings of that individual."
**Upload all plots, run parameter text files (file that has run settings saved), gain patterns of the two best and two worst individuals, etc**
|
|
|
163
|
Tue Jun 21 12:02:23 2022 |
Alex M | Run Details: Machtay_20201023_300K_Nus_50_Individuals | This run was started on 10/23/2020. The purpose was to attempt to demonstrate evolution by beginning from 50 identical individuals in the initial generation (which had previously produced a low score).
This run was the first time we removed the penalty on fitness scores for antennas which exceeded the borehole radius. It was also the first time we increased the number of neutrinos thrown up to 300,000. 50 fully asymmetric individuals were evolved over 25 generations.
There was a 50/50 split for roulette/tournament selection and 75%/10%/15% for crossover/reproduction/immigration. While the evolution was somewhat flat, we do believe we demonstrated evolution because the average score rose as the run evolved.
(Note that the final generation was interrupted but the plot was still made, hence the drop to 0 for all scores on the plot) |
| Attachment 1: Fitness_Scores_RG.png
|  |
|
|
168
|
Tue Jun 28 11:40:32 2022 |
Alex M | Run Details: Machtay_20200911_Symmetric | This was a symmetric run that began on 9/11/2020. This run used fewer neutrinos to determine how significant the number would be on the performance of the evolution. 30,000 neutrinos were used per individual.
10 individuals were evolved over 35 generations, using 100% roulette selection and the old mutation algorithm, where genes had a 40% chance of mutating. Mutations changed the gene by reselecting the value from a guassian with a mean and sigma equalt to the mean and sigma of that gene's value in the generation.
|
| Attachment 1: Fitness_Scores_RG.png
| 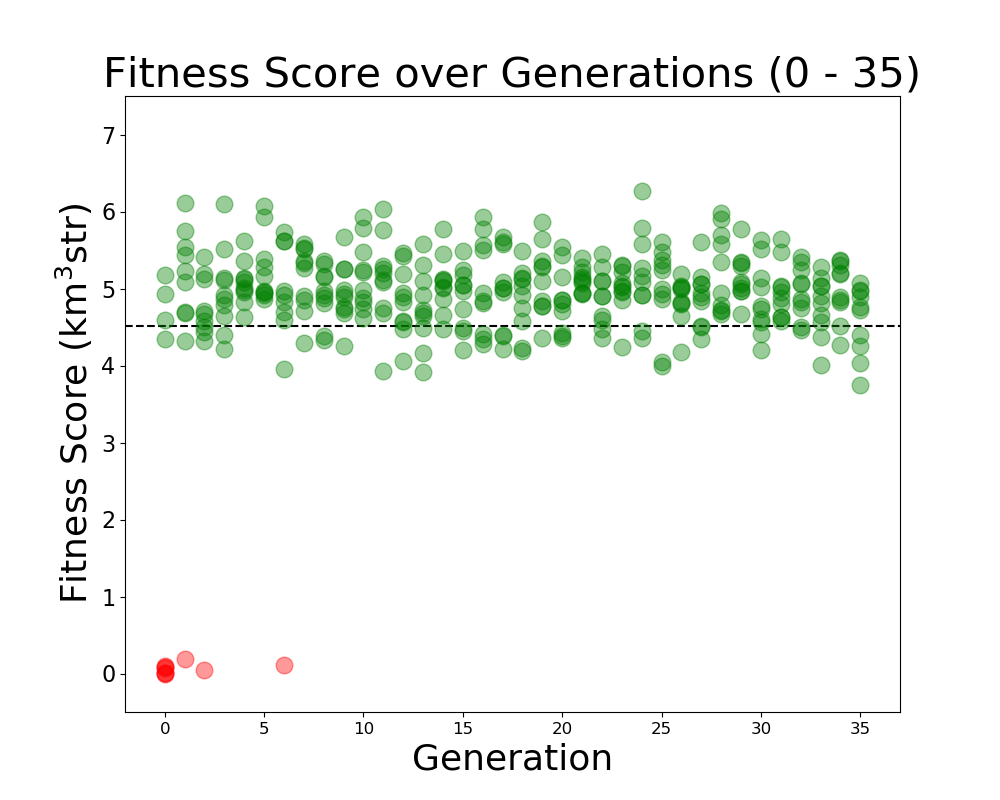 |
|
|
167
|
Tue Jun 28 11:35:15 2022 |
Alex M | Run Details: Machtay_20200831_Asym_Length_and_Angle | This run was conducted beginning on August 31, 2020.
Both the angle and length were asymmetric for the two cones. It was run with 10 individuals over 42 generations using 150,000 neutrinos. A penalty was still implemented on bicones exceeding the borehole width.
As this was before the substantial rewrite of the genetic algorithm, 100% of selection was done using roulette. Genes had a 40% chance to "mutate," which was done by selecting a new value from a guassian, with a mean and sigma equal to the mean and standard deviation of that gene's value in that generation.
|
| Attachment 1: Fitness_Scores_RG.png
| 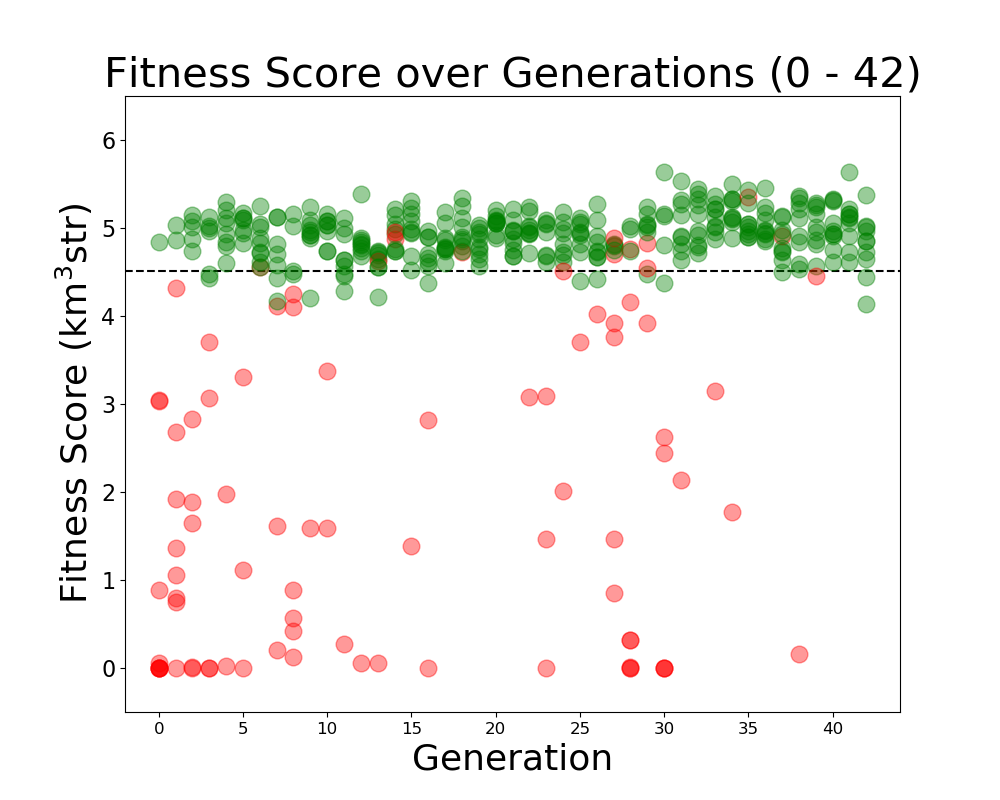 |
|
|
165
|
Tue Jun 21 12:22:02 2022 |
Alex M | Run Details: Machtay_20200827_Asym_Length_Run | This run followed the first symmetric run. It had an asymmetric length, but the opening angle and inner radius were kept symmetric.
10 individuals were evolved over 17 generations using 100,000 neutrinos. All selection was done through roulette, and all individuals were formed through crossover and the old mutation method (where individual genes had a 40% to be mutated, and mutated genes were chosen from a gaussian based on the mean and standard deviation of that gene's value in the generation). |
| Attachment 1: Veffectives_RG.png
| 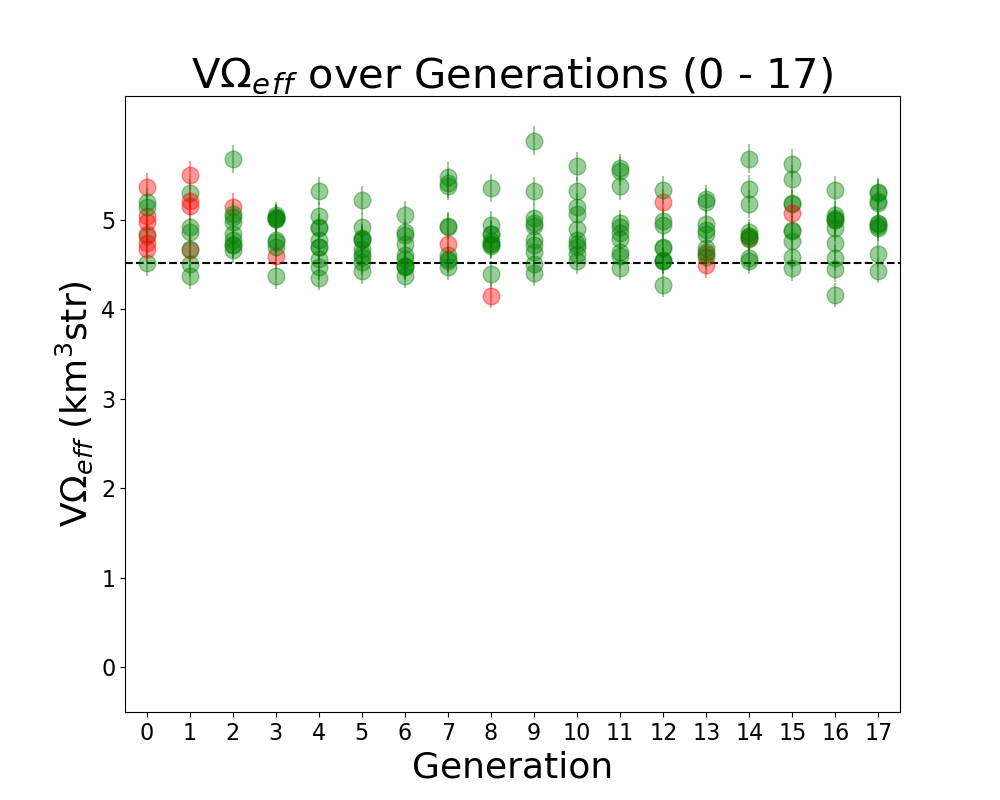 |
| Attachment 2: Fitness_Scores_RG.png
| 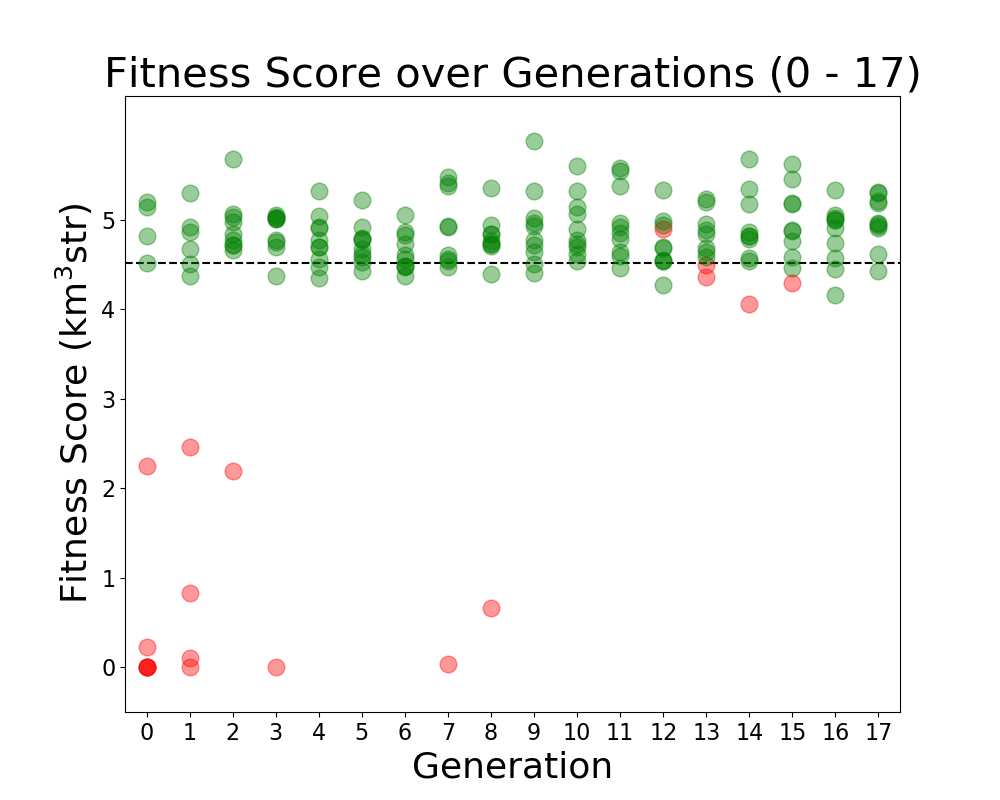 |
|
|
164
|
Tue Jun 21 12:15:18 2022 |
Alex M | Run Details: Machtay_20200824_Real_Run | This is considered to be the first run of "good" data. Prior to this run, we ran with few neutrinos and had errors which needed to be resolved in AraSim and with the way we handled XFdtd's simulation results being passed to AraSim. Those were resolved in the summer before this run.
This was a symmetric run of 10 individuals over 15 generations using 100,000 neutrinos per individual. This preceded the substantial modifications made by Ryan to the GA. All individuals were formed through crossover, but there was a probability of 40% for genes to be mutated. In this case, mutation was a complete change to the gene, though it was based on the average value of that gene in the generation. There was no reproduction or mutation. All individuals were selected through roulette.
Green individuals indicate that there was no penalty applied, while red individuals had a penalty factor multiplied by their effective volumes. The penalty was of the form exp(-(R-7.5)^2), where R is the outer radius, in cm, of the bicone. |
| Attachment 1: Fitness_Scores_RG.png
| 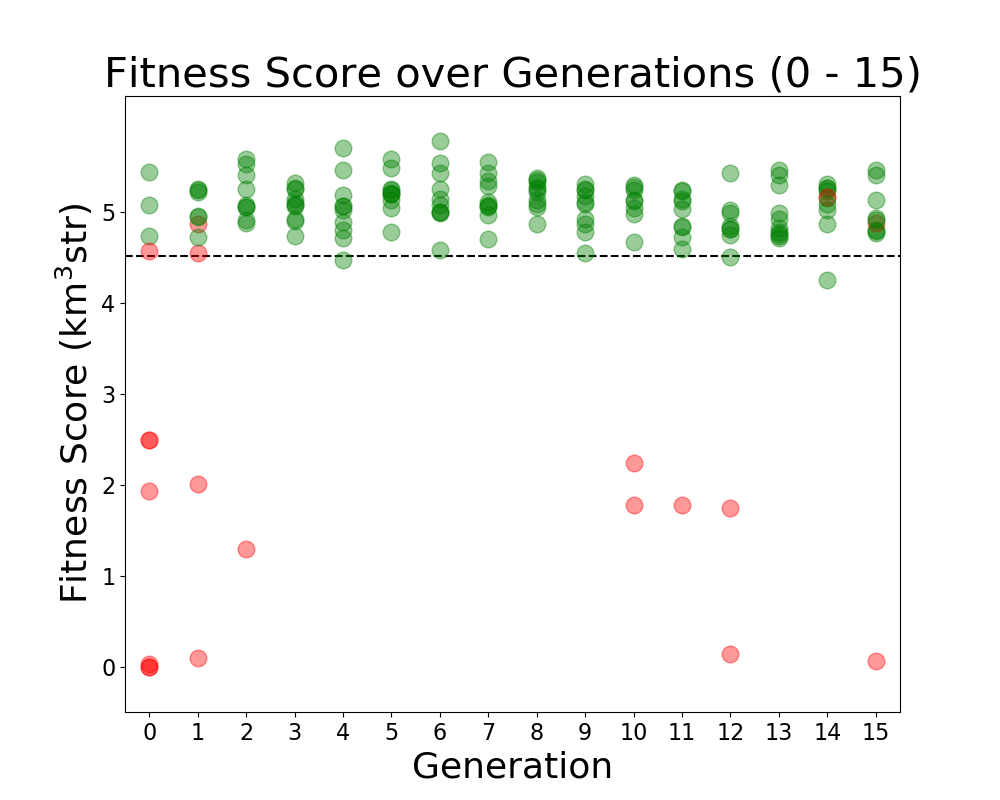 |
| Attachment 2: Veffectives_RG.png
| 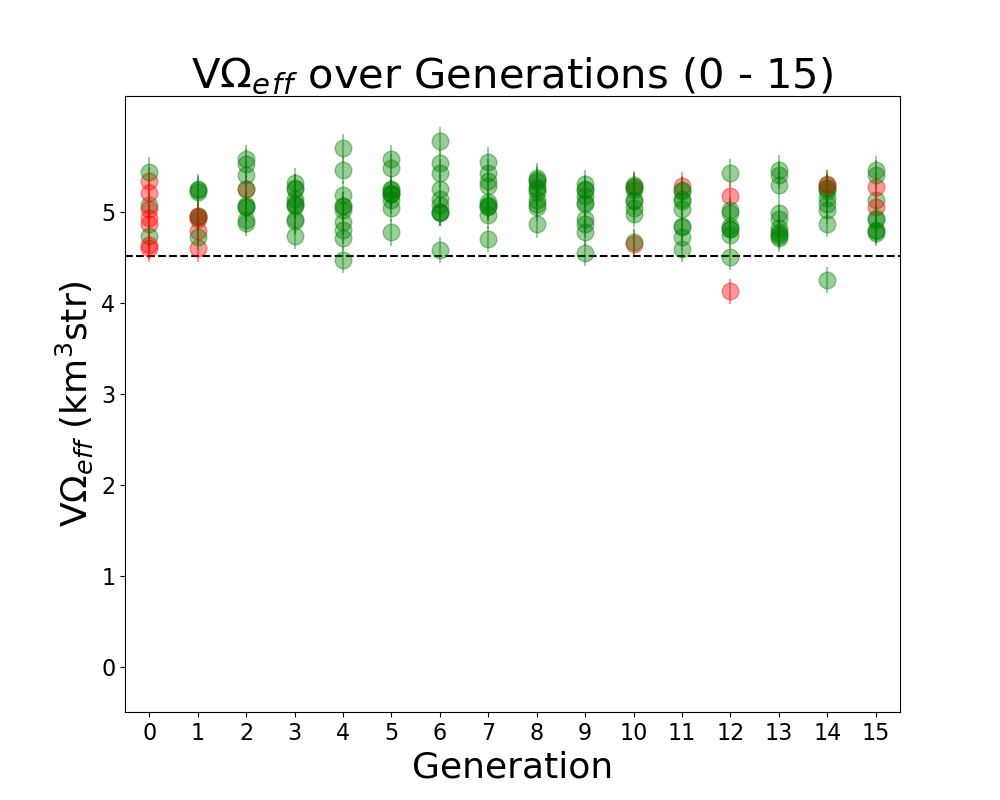 |
|
|
159
|
Tue Jun 7 11:29:48 2022 |
Alex M | Run Details: AraSim_Polarity_Fix_2021_03_19 | This is the run that is discussed in the GENETIS paper submitted to Physical Review D in December of 2021. It was conducted beginning on March 19, 2021. The preprint can be found here. It ran for 31 generations with 50 individuals per generation. Each individual was run through AraSim for 300,000 neutrinos.
The ratio of generative operators was: 72% crossover, 22% immigration, 6% reproduction.
The ratio of selection operators was: 80% roulette, 20% tournament, 0% rank.
The highest scoring individual had a fitness score of 5.24. However, when it was rerun with many more neutrinos (3*10^7), it had a score of 4.90, an 11% improvement over the ARA bicone.
This run was conducted before the introduction of the automatic run_details.txt generator. Instead, there is a file called Run_Notes.txt (attached) which details some of the errors that were encountered. These errors were remedied during runs by removing the offending AraOut file (which had too high of a fitness score due to a weight being calculated to be greater than 1, which shouldn't be possible) and have since been prevented by modifying the data file AraSim uses to calculate the weights. |
| Attachment 1: Run_Notes.txt
|
//Notes for AraSim_Polarity_Fix_2020_03_19 run
3/27/21: We ran into a rare error we see where AraSim gives an absurd weight (>>1) to a passed event. We reverted the state and replaced the effective volume in that file (AraOut_6_3_8.txt) with the average from the other AraOut files for that individual. DO NOT USE AraOut_6_3_8.txt WHEN ANALYZING ARASIM DATA.
5/9/21: We ran into the same error as above. We reverted the state and replaced the effective volume in the file (AraOut_23_7_7.txt). DO NOT USE AraOut_23_7_7.txt WHEN ANALYZING ARASIM DATA.
5/11/21: We ran into the same error. We are reverting the state and replacing the file (AraOut_25_10_6.txt). DO NOT USE AraOut_25_10_6.txt WHEN ANALYZING ARASIM DATA.
|
| Attachment 2: Violin_Plot_super_big.png
| 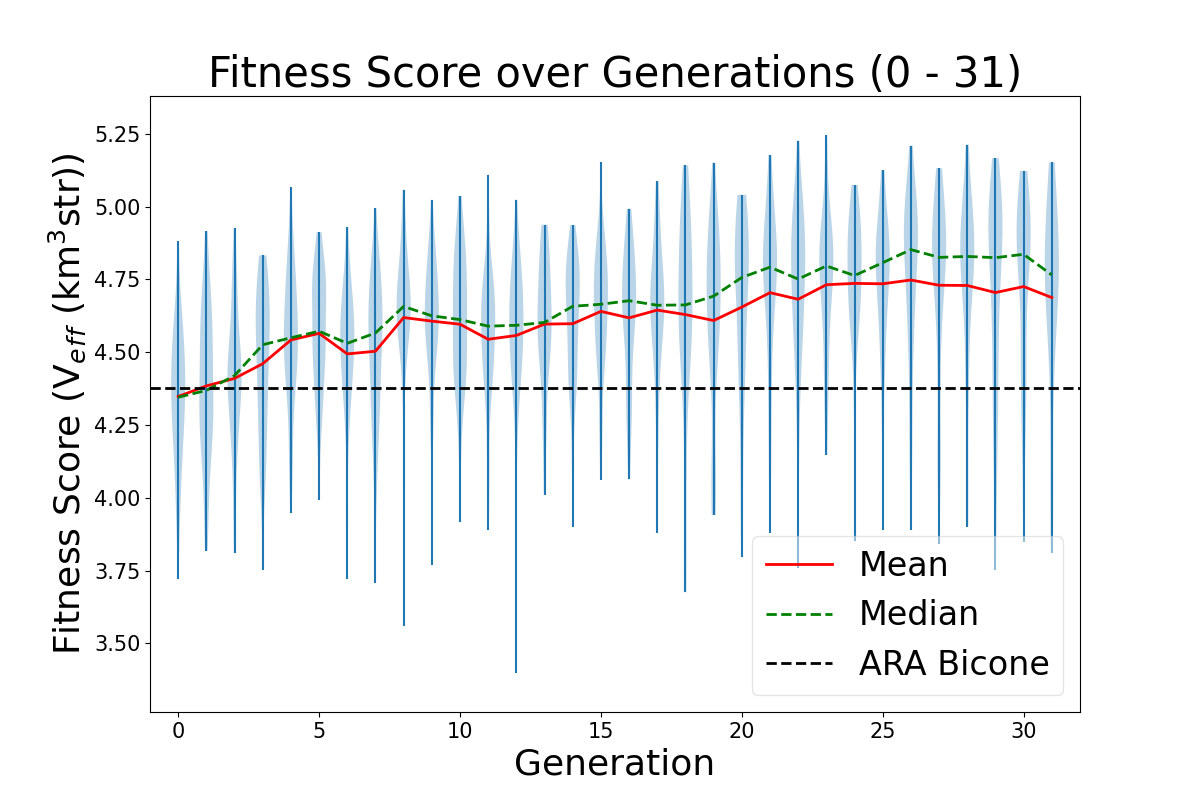 |
|
|
188
|
Fri Dec 30 01:16:52 2022 |
Alex M | Run Details: 2022_12_29 | We began a new run of the curved sided GA. Previously, we conducted a run just like this in July of 2022 (titled 2022_07_15_Latest_Greatest). In that run, we made substantial updates to the loop, including using the latest version of AraSim. However, we discovered after the run (which had a very flat fitness score growth curve) that we had not modified AraSim to take in gainfiles appropriately (so it was just evaluating the ARA bicone repeatedly). We've fixed this issue and a few more (see the github repository for the issues that exist and have been resolved) and are redoing this run. The run details will look much the same as that one and are visible in the attached file. Here is the location of the directory where the data for this run will be stored: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_12_29
For a list of changes made in this run compared to previous ones, see this ELOG post on the previous run: http://radiorm.physics.ohio-state.edu/elog/GENETIS/186 |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_12_29' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=100 ## number of individuals per generation; please keep this value below 99
Seeds=5 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=60000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=0 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=96 ## Number (not fraction!) of individuals formed through crossover
MUTATION=25 ## Probability of mutation (divided by 100)
SIGMA=6 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
174
|
Fri Jul 15 18:33:20 2022 |
Alex M | Run Details: 2022_07_15_Latest_Greatest | We have started a new run. Here is the directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_07_15_Latest_Greatest
This run is a substantial overhaul. It is a curved run using the latest parameters from Ryan's optimization loop. Here is a list of important changes:
- The newest version of AraSim has been implemented.
- The minimum length has been decreased to 10 cm per side.
- The GA is recording the parents and operators used to generate individuals.
- The number of individuals has been increased to 100 per generation.
- The number of neutrinos per individual has been increased to 600k.
This run may take longer than previous runs due to the increased number of individuals. There may need to be modifications made to resolve or work around AraSim errors that delay the loop (due to the auto-resubmit function). Find the details of the run attached. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_07_15_Latest_Greatest' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=100 ## number of individuals per generation; please keep this value below 99
Seeds=5 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=120000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=12 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=76 ## Number (not fraction!) of individuals formed through crossover
MUTATION=10 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
160
|
Tue Jun 7 12:20:23 2022 |
Alex M | Run Details: 2022_04_14_Identical_Asym_Lower_Min | This run was a short run (stopped due to resource limits) that was conducted to test the minimum length constraint that had been applied in the past. Previously, the minimum length was set to be 37.5 cm for each cone; in this run, it was lowered to 10 cm. It ran for 60 generations with 50 individuals per generation, each evaluated with 300,000 neutrinos. To compare to the paper run, this run was performed using the asymmetric algorithm, meaning each individual had 6 genes (length, inner radius, and opening angle for each cone). The initial generation began with identical individuals, with genes matching the genes of the best performing individual mentioned in the GENETIS paper.
The ratio of generative operators is as follows: 72% crossover, 22% immigration, 6% reproduction. Mutation was also used, with 1% of genes produced by crossover mutated by adding a number chosen from a Gaussian centered at 0 with a width of 5% of the gene's value.
The ratio of selection operators was: 20% roulette, 80% tournament, 0% rank, 0% elite.
Attached are the run details, violin plot, and "rainbow" plot. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_04_14_Identical_Asym_Lower_Min' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=0 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=8 ## Percent of individuals selected through tournament (divided by 10)
RANK=0 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
| Attachment 2: Violin_Plot.png
| 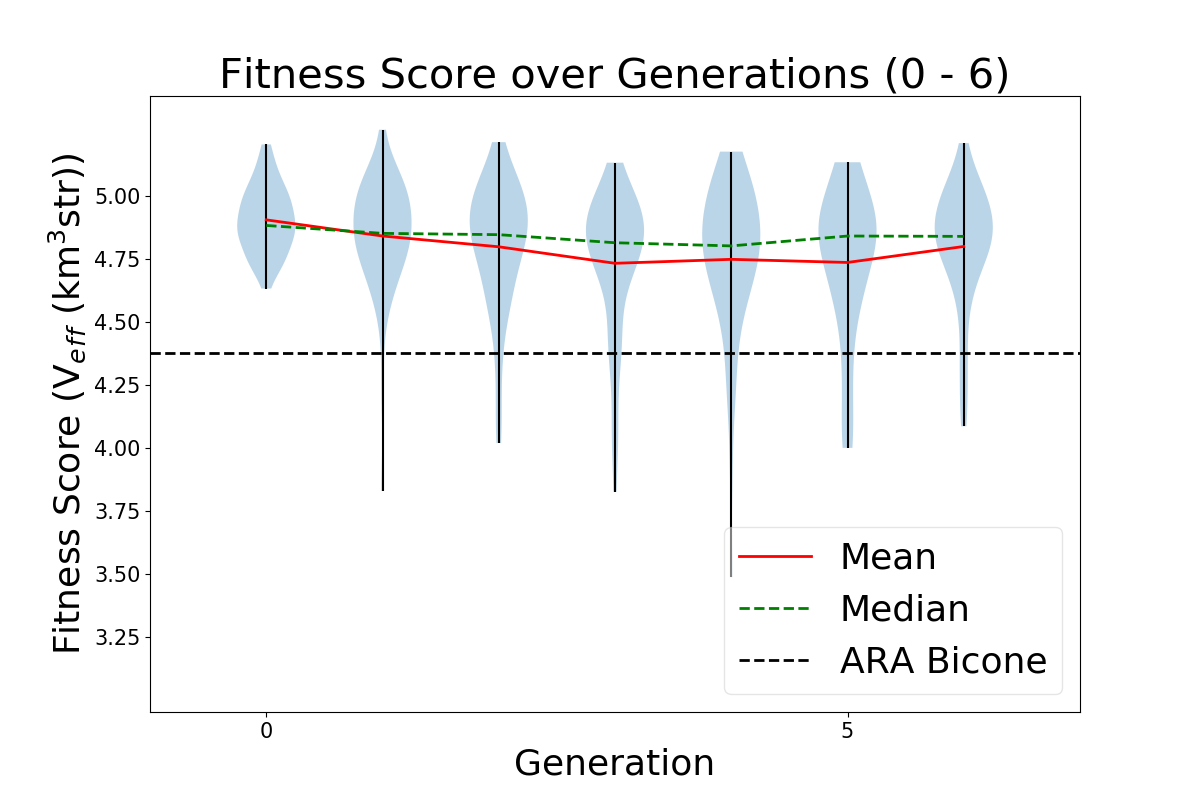 |
| Attachment 3: 2022_04_14_Identical_Asym_Lower_Min_Rainbow.png
| 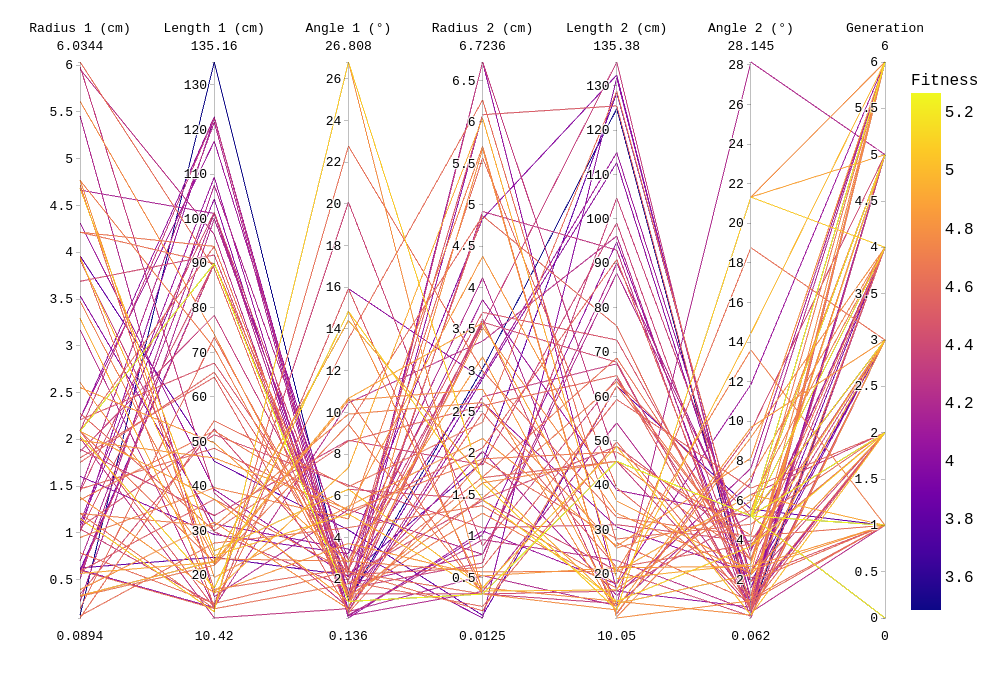 |
|
|
158
|
Tue Jun 7 11:01:59 2022 |
Alex M | Run Details: 2022_02_08_Rank_Test | This run was started on February 2, 2022. It is a full run of 50 generations with 50 individuals per generation using the quadratic version of the loop. This means that each individual is defined by 8 genes (length, inner radius, linear coefficient, and quadratic coefficient for each cone). Attached is the txt file run_details.txt that is automatically generated when the loop is run. Each individual was run for 300,000 neutrinos.
This run used the usual ratio of generative operators: 72% crossover, 22% immigration, and 6% reproduction. It also used the proper mutation function: 1% of genes created through crossover were mutated by adding a number chosen from a Gaussian distribution centered at 0 with a width of 5% of the gene's value (MUTATION WAS NOT USED BUT WAS AVAILABLE).
This run was the first full run in which the rank selection operator was used. The ratio of selection operators was: 0% roulette, 10% tournament, 90% rank, 0% elite.
This run used the script fitness_check.py to average the scores of identical individuals that appeared across multiple generations. This gives us a more accurate measure of those individuals' scores. It may also explain why this run appears so flat (on the violin plot): fluctuations in the scores should die down as repeated individuals have more accurate scores, and newly generated individuals that perform highly regress to their mean (actual) score when they are reproduced/recreated through crossover. This has led us to question our GA parameters, as Audrey and Autumn demonstrated that roughly 1/4 individuals in the run are repeat individuals. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_02_08_Rank_Test' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=0 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=1 ## Percent of individuals selected through tournament (divided by 10)
RANK=9 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
| Attachment 2: Violin_Plot.png
| 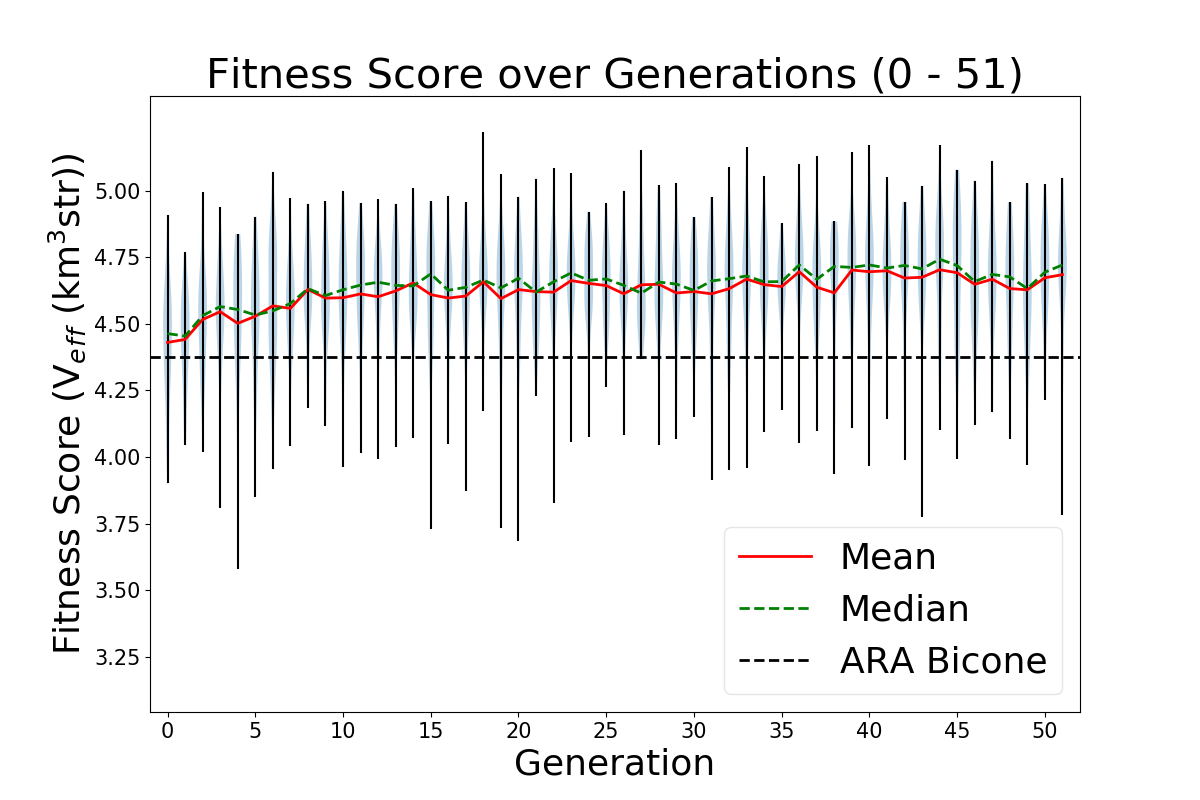 |
|
|
186
|
Mon Nov 28 10:44:27 2022 |
Alex M | Run Details Latest_Greatest_2022_11_26 | Here are the changes that have been made in this (and the preceding) run relative to what we have been doing before.
- The newest version of AraSim has been implemented.
- This was done because AraSim has gone through significant changes since 2019 that make it more accurate and makes it run faster.
- The minimum length has been decreased to 10 cm per side.
- This was done because we previously tested shorter lengths for a response to the reviewer when we submitting the paper to Phys Rev D. Previously, we thought that going below 37.5 cm would lead to unreliable results, but we are reassured that that is not the case now.
- The GA is recording the parents and operators used to generate individuals.
- This should allow us to look back and make a "history" of the evolution by seeing where genes come from in each generation.
- The number of individuals has been increased to 100 per generation.
- Ryan demonstrated that a higher number of individuals does lead to a substantial improvement in the speed at which the evolution converges.
- We previously thought that it did not make a big difference. However, that was done by measuring the average maximum fitness score. If we measure how many generations it takes for the evolution to reach a benchmark score, we find that more individuals helps significantly.
- The number of neutrinos per individual has been increased to 600k.
- This was done to make the fitness scores more precise. At 300k neutrinos, we expect an error of around 0.2. This will cut things down closer to 0.1, which may help us decrease the number of outlier measurements without increasing run time too much because of the AraSim speed up.
This run may take longer than previous runs due to the increased number of individuals. There may need to be modifications made to resolve or work around AraSim errors that delay the loop (due to the auto-resubmit function). Find the details of the run attached. |
| Attachment 1: run_details_2022_11_26.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_11_26_Latest_Greatest' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=100 ## number of individuals per generation; please keep this value below 99
Seeds=5 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=120000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=0 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=96 ## Number (not fraction!) of individuals formed through crossover
MUTATION=4 ## Probability of mutation (divided by 100)
SIGMA=6 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
201
|
Tue Mar 7 00:06:57 2023 |
Bryan Reynolds | Retroactive AREA update: January 18, 2022 AREA run- No linear freq. dependence test w/ increased NPop | The following is a run summary of an AREA test using 100 individuals per generation, without the linear frequency dependence on the gain, with the frequency fixed (i.e. the same gain pattern is produced for all frequencies). This run was a preliminary test of the AREA algorithm with a larger NPop (100 individuals per generation) after seeing promising results in the previous test that only used 20 individuals per generation. The percentages of selection methods/operators used attempted to mimic the optimal percentages used for the PAEA Bicone Loop, but because the algorithms do not set these in the same manner, these are highly unlikely to be the best percentages to use. The results show a very quick plateau in fitness score with a loss in diversity of solutions after a low number of generations, potentially meaning that a poor breakdown of selection methods/operators was chosen. This result seems to underscore the need for a test loop-style optimization for AREA to determine the best breakdown of selection methods and operators to use for AREA.
Run details:
- Run Type
- Run Date
- Run Name
- 20230118breynoldsrun_noLinearDependAREATest_38RCO_2RM_58TCO_2TM_150000NNU_2Seeds
- Why are we doing this run?
- This test increased the number of individuals to 100 and attempted to mimic the percentages of selection methods and operators found to be optimal for the PAEA loop.
- What is different about this run from the last?
- The previous test showed slow/minimal evolution after ~12 generations, and it was speculated that this was due to a small NPop of 20 individuals. This test uses an increased NPop of 100 individuals.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- 300,000 Total (150,000 NNU x 2 Seeds)
- Operatiors used (% of each):
- 96% Cross-Over, 4% Mutation
- Selection methods used (% of each):
- 40% Roullette, 60% Tournament
- Are we using the database?
Results:
- Summary and comments on results
- The fitness score quickly plaeaued and diversity of solutions was lost, potentially due to a unsuccessful choice of selection method and opterator percentages.
- An important future step is to work with Ryan to create a test loop optimization for the AREA algorithm, as it is written differently in how it is passed selection method and operator percentages and requires its own optimization to find the optimal ratios of these. (Work on creating a test loop optimization for AREA is currently underway).
EDIT 4/10/23
Re-uploading gain pattern associated with most fit individual from this run
|
| Attachment 1: 20230201_singleFreqTest_100NPop_fitnessPlot_10Gens.PNG
| 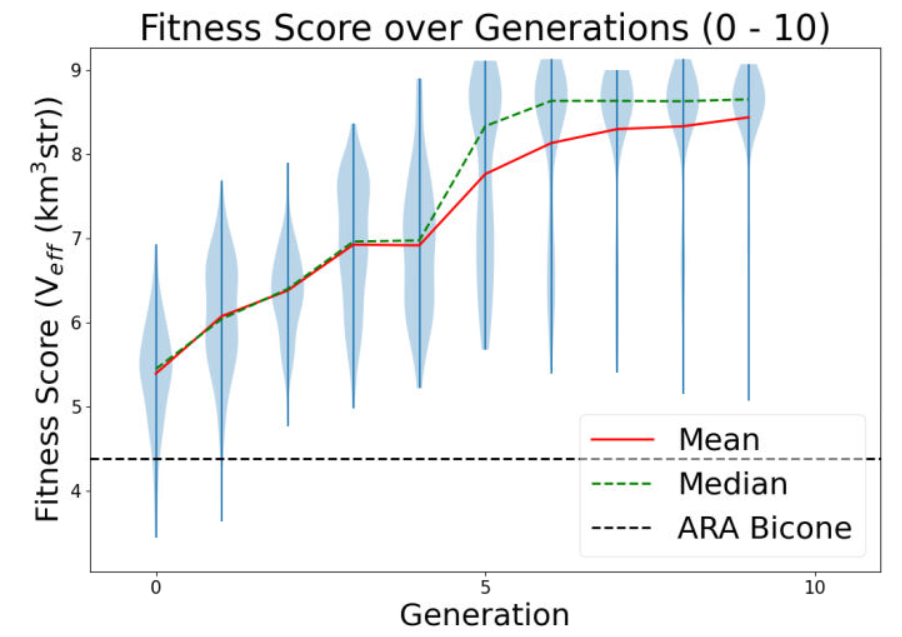 |
| Attachment 2: 20230410_AREA_mostFitIndivReUpload_singleFreqTest.PNG
| 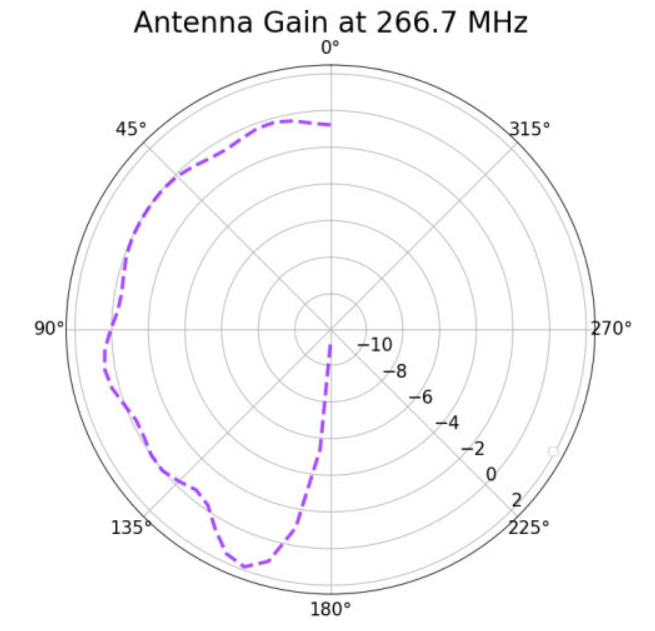 |
|
|
200
|
Mon Mar 6 23:25:12 2023 |
Bryan Reynolds | Retroactive AREA update: December 26, 2022 AREA run- No linear freq. dependence test | The following is a run summary of the AREA test without the linear frequency dependence on the gain, with the frequency fixed (i.e. the same gain pattern is produced for all frequencies). This run was a preliminary test of the AREA algorithm after fixing long-standing errors in the algorithm and finding that the initial constaints implemented did not work with the frequency dependence on gain. Results from this run did show a "proof-of-concept" that the AREA algorithm seems to evolve single frequencies in a promising way, however a low number of individuals may have stalled the evolution.
Run details:
- Run Type
- Run Date
- Run Name
- 20221226breynoldsrun_noLinearDependAREATest_8RCO_8RM_2TCO_2TM_30000NNU_10Seeds
- Why are we doing this run?
- We tested the AREA algorithm set to evolve at a single frequency in order to see if it would evolve a gain pattern at that frequency within the required constraints.
- What is different about this run from the last?
- This run only evolves gain patterns at a single frequency, allowing us to investigate the evolved gain patterns and check that the required constraints on the power conservation are met, without yet fixing the constraints function to work with the linear frequency dependence relationship. This is also the longest test run since the AREA algorithm was fixed and gotten into working order.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- 300,000 Total (30,000 NNU x 10 Seeds)
- Operatiors used (% of each):
- 50% Cross-Over, 50% Mutation
- Selection methods used (% of each):
- 80% Roullette, 20% Tournament
- Are we using the database?
Results:
- Summary and comments on results
- The evolution appeared to be successful in that the most fit gain pattern found (attached) seems to make sense with our understanding of the underlying physics. Additionally, after inspecting the outputs, conservation of power was confirmed to be properly constrained. However, in the attached violin plot, the fitness score plateaued quickly and never evolved further, with the best individual coming in generation 12. This was speculated to be due to the small population used in this test (only 20 total individuals per generation).
|
| Attachment 1: 20230109_singleFreqTest_fitnessPlot_update30Gens.PNG
| 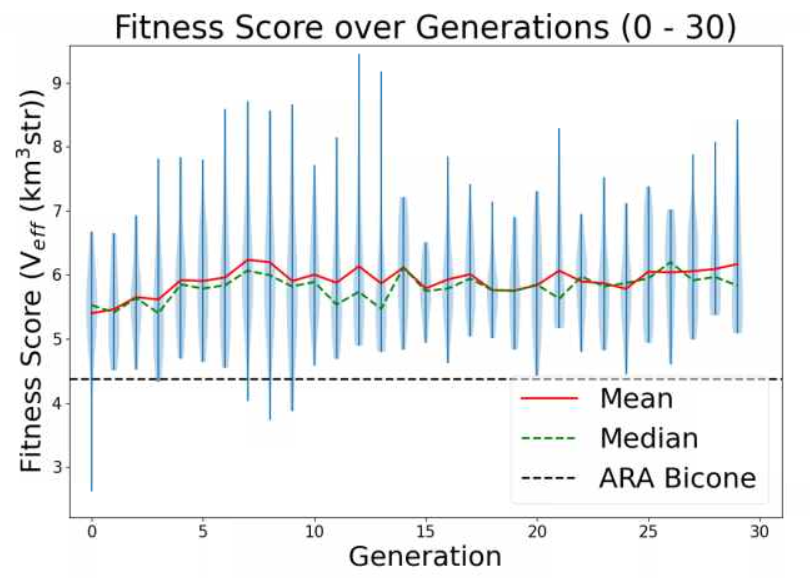 |
| Attachment 2: 20221229_singleFreqTest_overallBestIndiv_gen12Indiv17.PNG
| 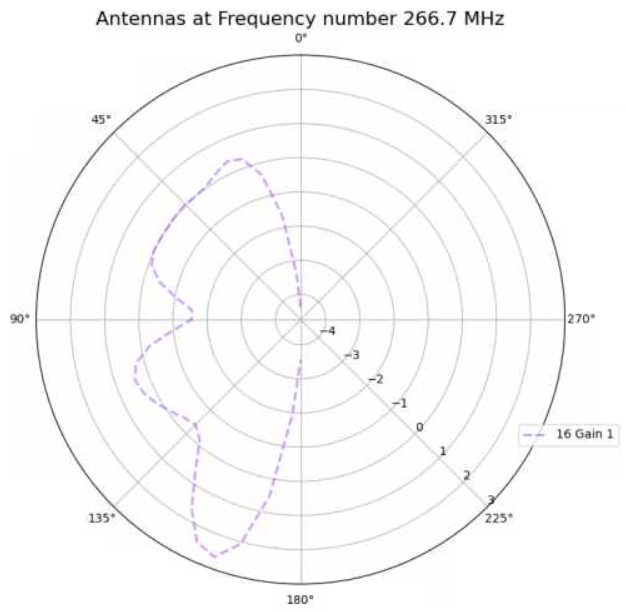 |
|
|
213
|
Thu Apr 27 01:36:32 2023 |
Alex M | Results from paper symmetric run | The reviewer asked us to run the loop again but for just symmetric antenna designs. We have completed that run and have results ready. We still need to resimulate the top five individuals, which I'll update this post with later. I attached the violin plot (though it should be fixed to not let the legend overlap the data). See this previous entry for the details of the run: http://radiorm.physics.ohio-state.edu/elog/GENETIS/198 |
| Attachment 1: Violin_Plot.png
| 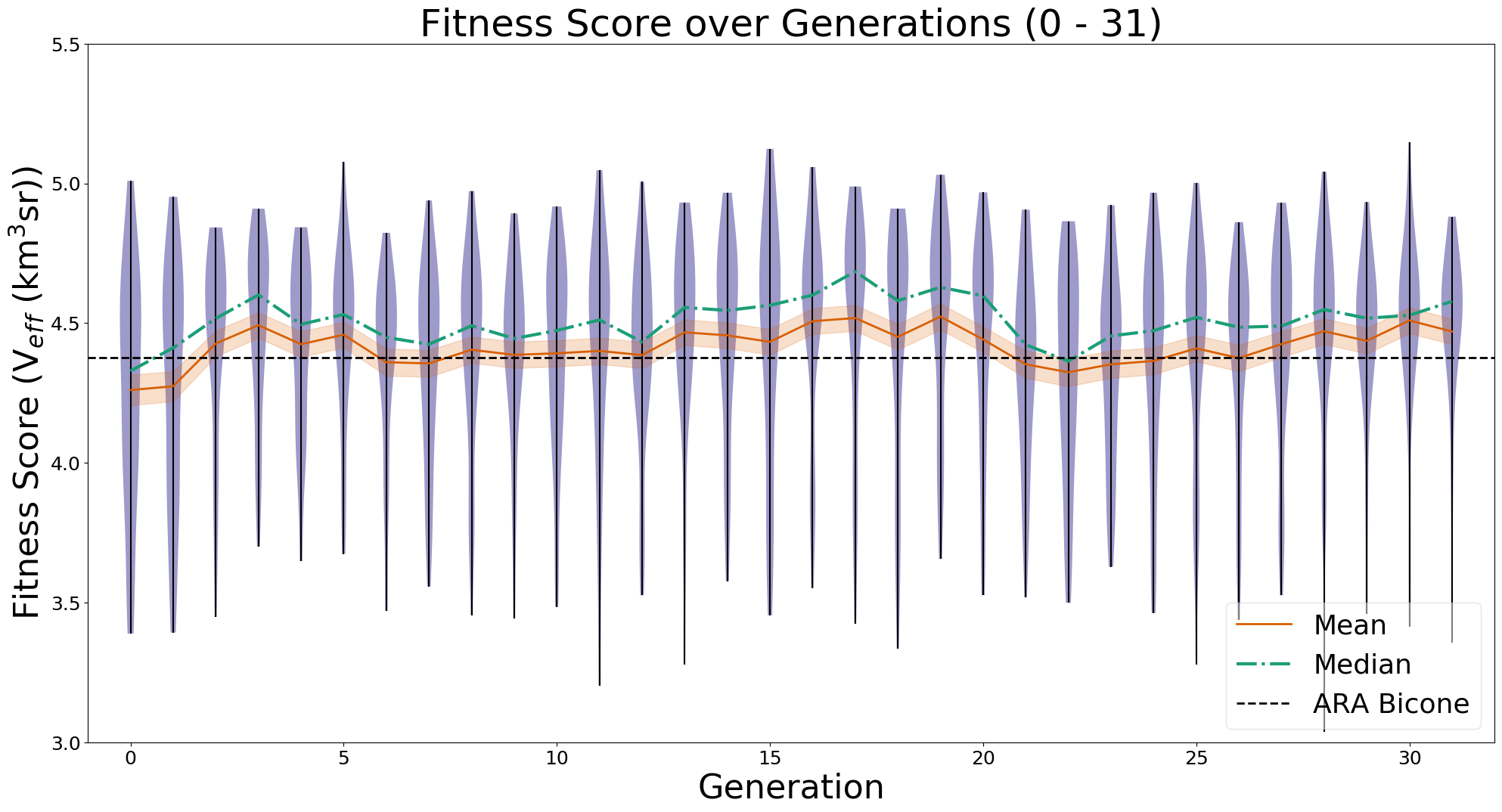 |
|
|
24
|
Mon Dec 2 16:41:35 2019 |
Julie Rolla | Requesting an interactive job | Requesting an interactive job:
qsub -I -X -l walltime=1:00:00 -l nodes=1:ppn=40
(ppn=24 makes it quicker FYI)
Note that walltime is the length of time you'll need it. You will likely need it for 4+ hours if you are running the loop... Probably longer. Note that before you enter this command into your command line, you must be logged into OSC.
For submitting a job with GPUs (which we will be doing to speed up XF now as of 1/24/20), use this command:
qsub -I -X -l walltime=1:00:00 -l nodes=1:ppn=40:gpus=1:default OUTDATED
srun -A PAS0654 -t 1:00:00 -N 1 -n 40 CURRENT |
|
|
144
|
Tue Feb 8 15:56:42 2022 |
Machtay | Rank Test Run | I fixed a bug in the loop, so we started another rank test run. Run details in the attached file.
The bug was searching for the generationDNA.csv file in the wrong place, meaning that it wasn't able to copy it to the run directory. That meant we didn't have a record of the generation data in the usual format. I don't think that this explains the flatness, since the the generationDNA.csv file was still created every generation correctly, so the GA knew where it was. But this test now corrects that problem and tests the usage of the rank selection method. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_02_08_Rank_Test' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=0 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=1 ## Percent of individuals selected through tournament (divided by 10)
RANK=9 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
Draft
|
Mon May 29 20:21:07 2023 |
Dylan Wells | Pueo Physics of Results Plots | The Physics of Results Plots have been added to the Pueo Loop. The current version of the plotter is built for pueoSim v1.0 and located in ${WorkingDir}/Antenna_Performance_Metric (Hasn't been pulled into the loop directory yet).
The pueoSim v1.0 IceFinal files were missing information on the RF direction and information needed to see an amplitude spectrum. I asked Will, and he said that the new version of pueoSim (v1.1.0) outputs the needed information.
I created an updated version of the plotter, and have a pull request here: https://github.com/osu-particle-astrophysics/GENETIS_PUEO/pull/40
However, before this can be implemented in the loop, we need a way to get errors from pueoSim v1.1.0.
Currently, there is a version of rootAnalysis.py here that can analyze the new root files and output fitness scores and errors, but instead of taking 20 minutes for a generation, it now takes 20 minutes per individual to run.
This is because the update splits the IceFinal file into IceFinal_allTree, IceFinal_passTree0, and IceFinal_passTree1. IceFinal_allTree and IceFinal_passTree1 are of comparable size to the previous IceFinal file, but the passTree0 final is
about 10-20 times larger, causing a slowdown in calculations. If we calculate without this file, it takes about 1 minute per individual, still a bit slower than before.
Ideas to solve this issue:
Don't use the passTree0 files (I have asked Will what they're for, and if we need them. Hopefully he responds after the Holiday.)
Instead of running through a Python for loop, call a C++ script that creates a CSV file the Python program can easily load in.
Submit a batch job to run the analysis in parallel before combining the outputs in the correct format. (We should maybe do this anyways?)
Continue using pueoSim v1.0 (I think we can still retain the Theta graphs for the incoming neutrinos, but there won't be any RF graphs or the possibility of amplitude spectrum plots)
Attached is a preview of plots the v1.1 plotter is capable of making.
Add weights.
|
| Attachment 1: 1_neutrinoFlavor_bestindiv.png
|  |
| Attachment 2: 1_nuDirCosTheta_bestindiv.png
| 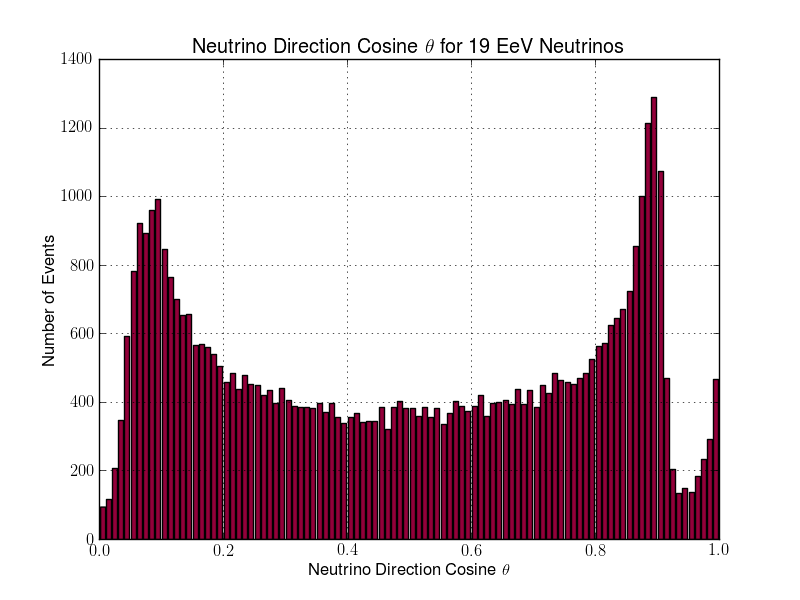 |
| Attachment 3: 1_nuDirTheta_bestindiv.png
| 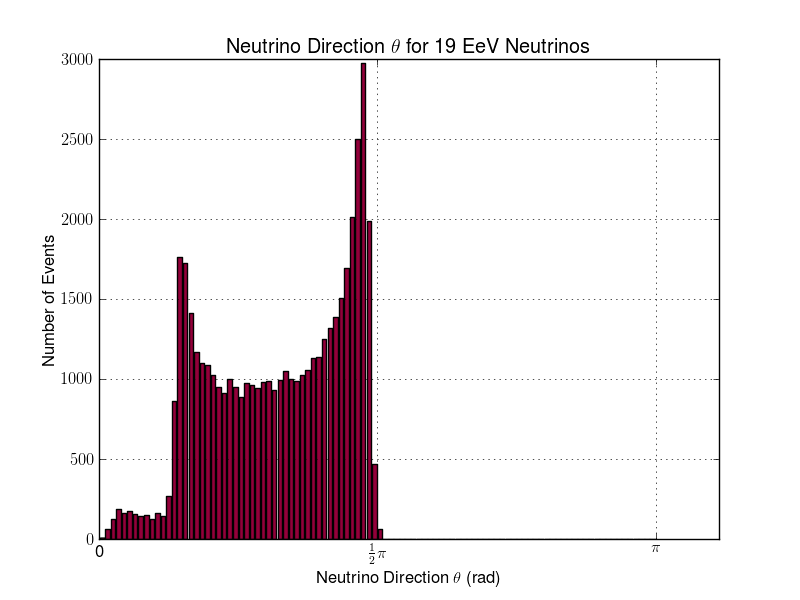 |
| Attachment 4: 1_RFdirCosTheta_bestindiv.png
| 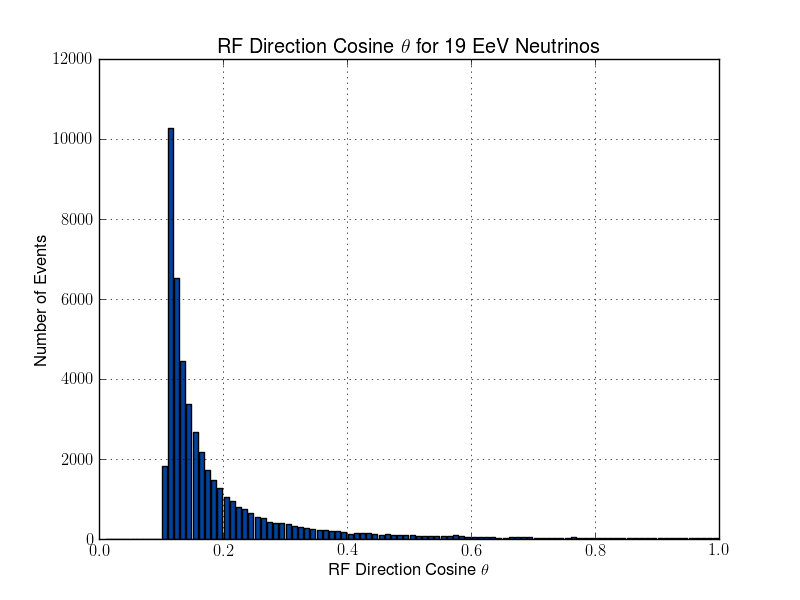 |
| Attachment 5: 1_RFdirTheta_bestindiv.png
| 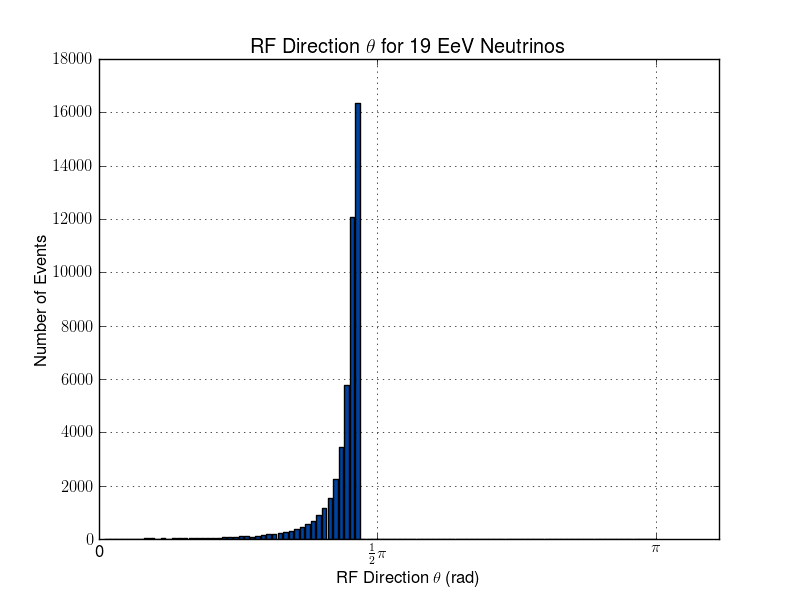 |
| Attachment 6: 1_maxEField_vs_maxEFieldFreq_2d_bestindiv.png
| 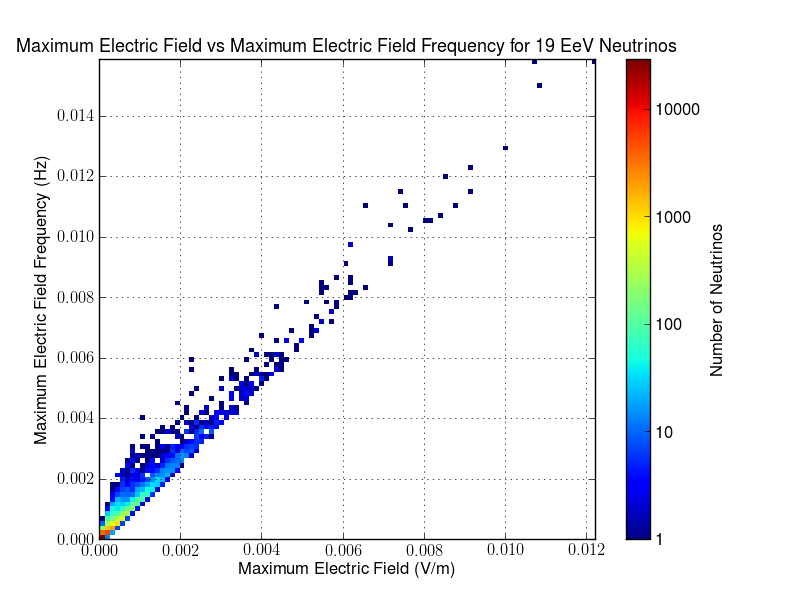 |
| Attachment 7: 1_1_signals_bestindiv.png
| 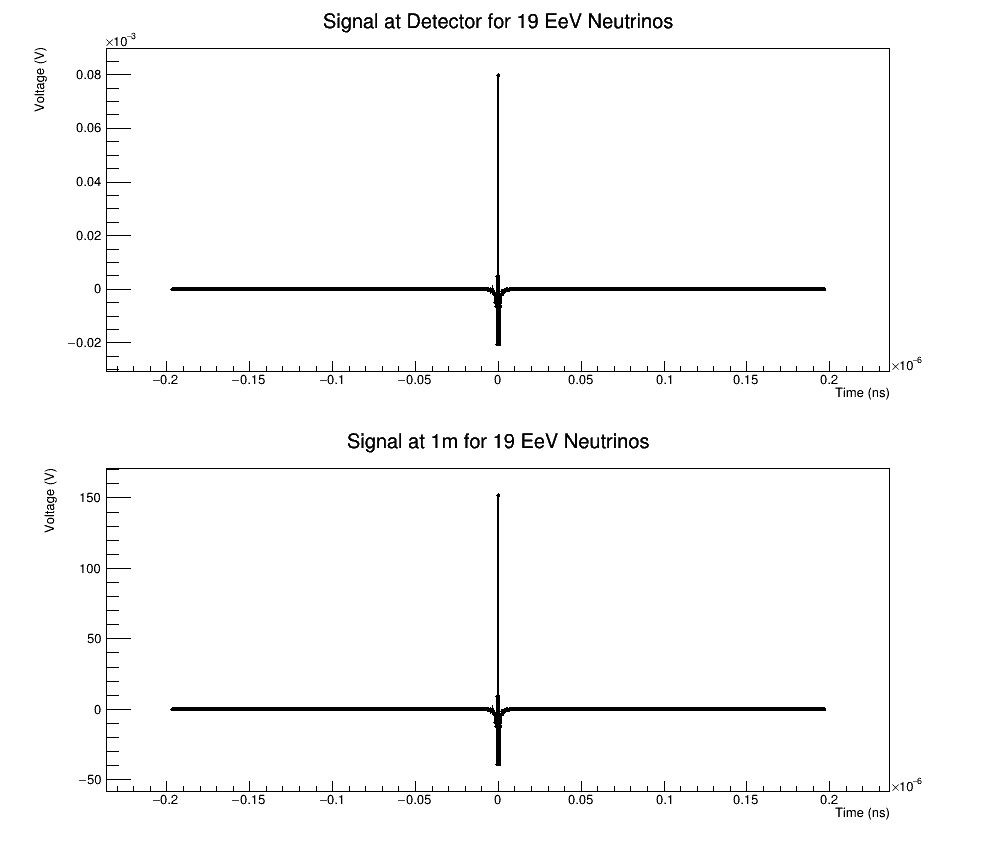 |
|
|
6
|
Fri Feb 22 15:43:36 2019 |
Julie Rolla | Proposal requests for XF (Remcom) | Here are a list of things we wish XF would do. We are keeping this list in hopes to add it to a proposal with them.
- Save pictures of CAD drawings of simulations.
- Close/quit XF GUI. App.quit doesn't actually work for us.
- Suppress the GUI and run solely through terminal.
|
|
|
214
|
Sun Apr 30 03:45:07 2023 |
Alex M | Preliminary PUEO Run | We have the PUEO loop in working order, though there are a few errors in accuracy and efficiency. We started a preliminary run with few individuals and neutrrinos as a large scale test that may still give us some useful data.
For this run, we are only evolving the inner side length (10cm to 25 cm) and height (10 cm to 50 cm). The walls are slanted at 45 degrees, so the outer length is completely defined by those two. The ridges are kept at 6 cm wide and 4 cm from the walls at the base. The curvature is set to 0,1 and the ridges are set to have the same height as the walls.
At the moment, we are using 24 individuals and 300,000 neutrinos. I'll update this ELOG post with more details from the run as they emerge. |
| Attachment 1: PUEO_run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_04_29_Test_10' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=24 ## number of individuals per generation; please keep this value below 99
Seeds=2 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=1000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
PUEO=1 ## IF 1, we evolve the PUEO quad-ridged horn antenna, if 0, we evolve the Bicone
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=6 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=10 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
|
|
|
150
|
Fri Apr 1 16:35:50 2022 |
Ryan Debolt | Population test. | https://docs.google.com/spreadsheets/d/1vvcmjByKfcns0-tbAjtePB8ZVGsXAKxXCfbMc99weMI/edit?usp=sharing Here is the spreadsheet link for the population test. |
|
|
Draft
|
Fri Sep 3 14:06:39 2021 |
Alex M | Plots for 9/3/21 Collaboration Meeting | Here are plots I made for the meeting on 9/3/21. These plots represent a comparison of the gain and realized gain for the 23rd generation of the run being discussed in the upcoming paper. Here is a list of the plots
- Gain vs realized gain
- Polar plots of the best individual from generation 23
- S11/VSWR plots
- Shows the S11/VSWR over the bandwidth for the best individual
|
|
|
136
|
Fri Sep 3 14:28:55 2021 |
Alex M | Plots for 9/3/21 Collaboration Meeting | This ELOG post contains plots I made this week for comparing the antennas as they were evolved in the run being discussed in the upcoming paper with those same antennas when using realized gain instead of gain. These plots are preliminary, in that they should be edited before being placed in a paper (for example, VSWR is not in dBi).
Plots:
- Gain vs Realized gain
- Polar plot showing the gain and realized gain of the best individual from the run in the paper
- VSWR/S11
- The VSWR of the best individual in the run over the frequency bandwidth
- Gain differences
- The difference between the gain and the realized gain for the best individual over the frequency bandwidth
|
| Attachment 1: polar_plot_300.04.png
| 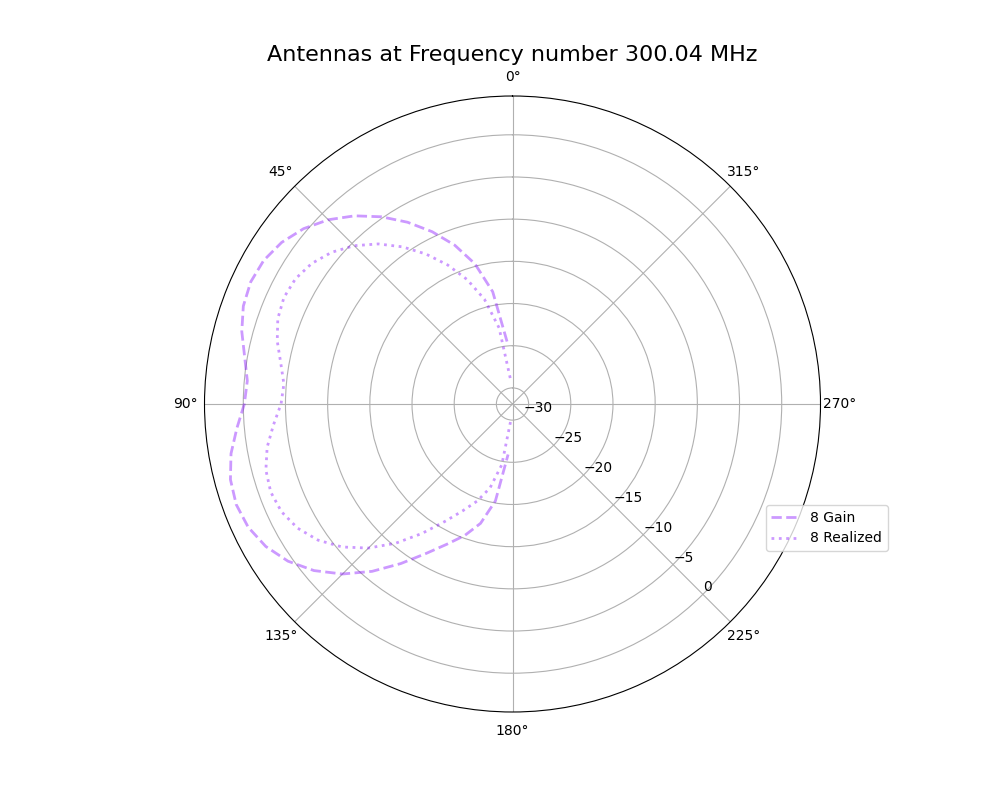 |
| Attachment 2: polar_plot_200.02.png
| 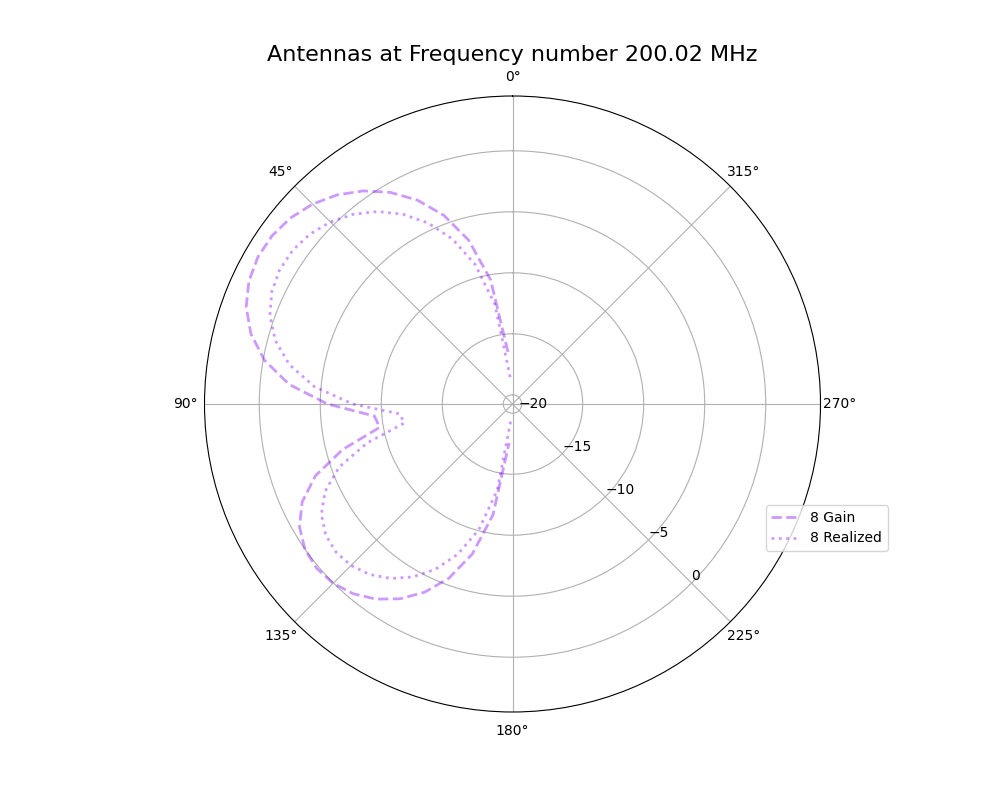 |
| Attachment 3: VSWR_plot_1811.png
| 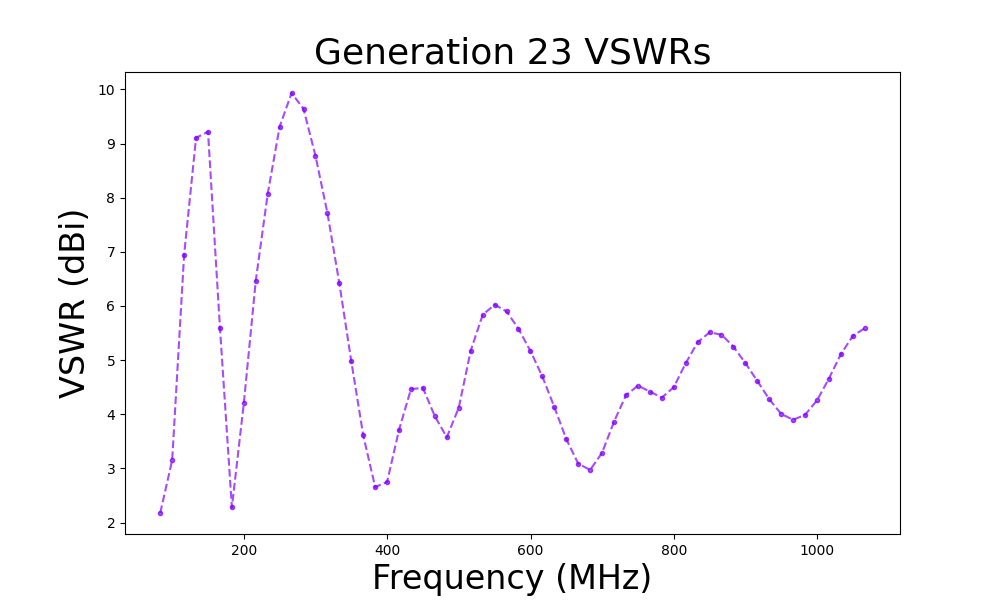 |
| Attachment 4: mean_gain_difference_1811.png
| 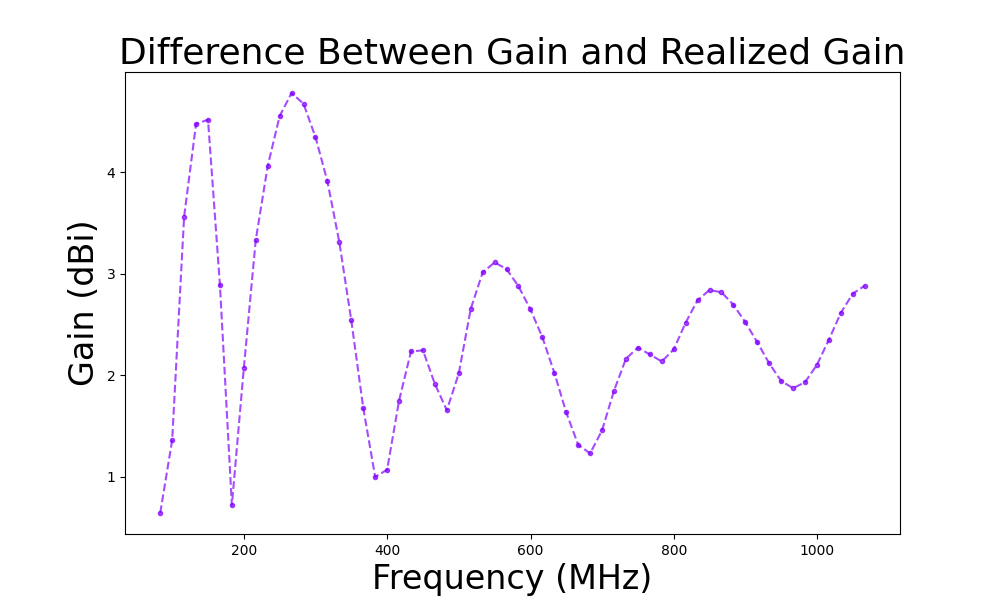 |
|
|
196
|
Sun Feb 19 17:12:34 2023 |
Jack Tillman | Physics Plots - 9_50, 13_84, 18_89, 19_96, 29_87 | Attached is a pdf of physics plots generated for the 9_50, 13_84, 18_89, 19_96, and 29_87 crazy sides individuals along with another containing information on the amount of triggered events and effective volumes for each individual. The crazy sides individuals were simulated for 3 million events using the same Arasim version as is currently in the loop. |
| Attachment 1: Feb2023_Physics_Plots_Powerpoint.pdf
|
| Attachment 2: Crazy_Sides_Triggered_Event_Data.pdf
|
|
|
192
|
Mon Feb 6 13:20:26 2023 |
Dylan Wells | Physical Paramaters for the best antenna | Asym Straight Sides (from the paper - Generation 23, Individual 8)
(inner radius, length, opening angle in radians)
2.08711,89.924,0.0161734
0.30175,45.3616,0.0910478
individual found in
/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/AraSim_Polarity_Fix_2021_03_19/AraSim_Polarity_Fix_2021_03_19.xf/Simulations/001108/Run0001/
|
|
|
145
|
Fri Feb 11 16:09:24 2022 |
Ryan Debolt | Parents.csv | Below is an example of our Parents.csv file written by the GA. This file tracks the parents of the individuals of the current generation. The columns and their contained information are as follows:
Current Gen:
The numbered individual of the current generation.
Parent 1:
The number of the first parent of this individual as read from the previous generation.
Parent 2:
The number of the first parent of this individual as read from the previous generation.
Operator:
The genetic operator that created the individual in that row. |
| Attachment 1: Parents.csv
|
Location of individuals used to make this generation:
Current Gen, Parent 1, Parent 2, Operator
1, 15, NA , Reproduction
2, 22, NA , Reproduction
3, 35, NA , Reproduction
4, 34, 39, Crossover
5, 34, 39, Crossover
6, 35, 29, Crossover
7, 35, 29, Crossover
8, 37, 5, Crossover
9, 37, 5, Crossover
10, 28, 12, Crossover
11, 28, 12, Crossover
12, 5, 22, Crossover
13, 5, 22, Crossover
14, 18, 23, Crossover
15, 18, 23, Crossover
16, 3, 33, Crossover
17, 3, 33, Crossover
18, 28, 14, Crossover
19, 28, 14, Crossover
20, 2, 17, Crossover
21, 2, 17, Crossover
22, 23, 22, Crossover
23, 23, 22, Crossover
24, 13, 15, Crossover
25, 13, 15, Crossover
26, 31, 35, Crossover
27, 31, 35, Crossover
28, 42, 13, Crossover
29, 42, 13, Crossover
30, 17, 6, Crossover
31, 17, 6, Crossover
32, 1, 5, Crossover
33, 1, 5, Crossover
34, 29, 38, Crossover
35, 29, 38, Crossover
36, 19, 10, Crossover
37, 19, 10, Crossover
38, 9, 38, Crossover
39, 9, 38, Crossover
40, NA, NA, Immigration
41, NA, NA, Immigration
42, NA, NA, Immigration
43, NA, NA, Immigration
44, NA, NA, Immigration
45, NA, NA, Immigration
46, NA, NA, Immigration
47, NA, NA, Immigration
48, NA, NA, Immigration
49, NA, NA, Immigration
50, NA, NA, Immigration
|
|
|
147
|
Thu Feb 24 20:08:46 2022 |
Ryan Debolt | Parents | |
| Attachment 1: Parent_tracking_example.pdf
|
|
|
232
|
Tue Jul 18 22:04:22 2023 |
Dylan Wells | Parallelizing pueoSim jobs | Currently, the PUEO loop runs pueoSim with 1 pueoSim process per job submitted.
Each of these jobs has 1 node and 8 cores, however, pueoSim only needs a single core to run.
Here is some data I collected by running the same seed of pueoSim with different numbers of cores in the job:
| Processes |
Cores Per Job |
Run Time (1000 neutrinos) |
Neutrinos Per Second |
| 1 |
1 |
100 |
10 |
| 1 |
8 |
102 |
10 |
| 1 |
16 |
96 |
10 |
| 1 |
40 |
96 |
10 |
So, we should be able to run multiple processes of pueoSim without losing much performance.
Tests with more cores:
| Processes |
Cores per Job |
Run Time (1000 neutrinos) |
Neutrinos Per Second |
| 8 |
8 |
106 |
75 |
| 8 |
16 |
126 |
63 |
| 8 |
40 |
107 |
75 |
| 20 |
20 |
142 |
140 |
| 20 |
40 |
138 |
144 |
| 32 |
40 |
178 |
179 |
| 40 |
40 |
208 |
192 |
A test comparing the parallelization with the 40000 neutrinos per process we simulate in the loop:
| Processes |
Cores Per Job |
Run Time (40000 neutrinos) |
Neutrinos Per Second |
| 1 |
8 |
3226 |
12 |
| 40 |
40 |
6651 |
240 |
So, these tests suggest we can simulate neutrinos ~20 times faster than currently done in the loop by utilizing more cores per job.
This method of multiple processes per job should be applicable to how we run other simulation software like AraSim as well.
(Note: both of these jobs had comparable queue times of 27 and 31 seconds respectively.
Also, I have tested and seen that the outputted root files are identical for the same seed regardless of if ran with multiple processes at the same time or with processes in serial)
(data found in /users/PAS1960/dylanwells1629/buildingPueoSim/testingouts/times.txt)
How to run multiple processes of pueoSim:
Previous Call:
./pueoBuilder/build/components/pueoSim/simulatePueo -i pueo.conf -o ${PSIMDIR}/outputs/${gen}_outputs/$SLURM_ARRAY_TASK_ID -r $run_num -n $NNT -e $Exp > $TMPDIR/out.txt
Multiple Processes Test Call:
for ((i=0; i<$threads; i++))
do
./simulatePueo -i pueo.conf -o $TMPDIR -r $i -n $NNU -e 19 > pueo.out &
done
wait
Instead of just calling peuoSim once per job, we can use the & symbol and a loop to run $threads number of pueoSim processes in the background and then wait for them to finish. |
|
|
234
|
Tue Jul 25 16:07:25 2023 |
Dylan Wells | Parallelizing XF and pueoSim in the loop | Standard Loop Architecture:
Complete an evolutionary step FOR EACH antenna before continuing on with the next step.
Steps:
1. Run the Genetic Algorithm for the entire population.
2. Run the XF radio simulation for the entire population.
3. Run the neutrino simulation software for the entire population.
4. Run root analysis and plots for the entire population.
However, due to constraints on the amount XF keys we have, we can only run 4-5 XF simulations at a time.
So, when the first antennas finish their XF simulations, their outputs will simply sit there until EVERY other antenna finished their XF simulations.
New Proposed Architecture:
Complete necessary evolutionary steps as a population, but string together those that don't rely on data from other antennas.
Steps:
1. Run the Genetic Algorithm for the entire population
2. Run the XF radio simulation for each antenna
- When an XF simulation finishes for an antenna, submit the pueoSim / root analysis jobs for that antenna
3. Run the plots for the entire population
This will allow us to complete most of our pueoSim computation while the XF portion of the loop is still running, cutting down the time between the final XF job finishing and the final pueoSim job finishing.
Test run of this new architecture with 100 antennas, 7,840,000 neutrinos per antenna.
| Part |
Time (seconds) |
| A |
1 |
| B1 |
1347 |
| Entire B2 |
54017 |
| B2 XF |
53063 |
| B2 remaining PUEO |
954 |
| E |
7 |
| F |
23 |
After the final XF job finished, the pueoSim simulations and analysis of outputted root files were completed in 954 seconds,
Comparison Between the Two Architectures (both using job optimization from Elog 232)
| Architecture |
Neutrinos |
Time In Loop (s) |
| Standard |
4,000,000 |
~6500 |
| New Proposed |
7,840,000 |
~950 |
Notes on Chart:
Source of data located in /users/PAS1960/dylanwells1629/buildingPueoSim/testingouts/times.txt and /fs/ess/PAS1960/HornEvolutionTestingOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_07_24_test5/time.txt respectively)
Time from standard uses the time one of the 250 jobs running in parallel took in my testing of parallelizing processes inside of pueoSim jobs: 250 jobs * (40 * 40000 neutrinos per job) / 100 individuals = 4,000,000 neutrinos per individual)
The New Proposed time includes time spent analyzing the outputted root files to find fitness scores and errors, which would have taken around 100 seconds * population size for its number of neutrinos and files per individual (/fs/ess/PAS1960/HornEvolutionTestingOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_07_23_test5/time.txt for data on this number)
So, this new architecture can provide more improvements for the amount and speed of neutrino simulation in the loop on top of the methods discussed in Elog 232.
This architecture could also be applied to see improvements in the Bicone and Hpol loops which are both affected by the limited number of XF keys.
Additional Notes:
1. For this new architecture test, each antenna uses 49 jobs for neutrino simulation instead of 2.5 previously. (49 pueoSim + 1 XF for 50 jobs per antenna, 5,000 jobs per generation)
2. The time for each antenna to submit, queue, and finish its neutrino simulation jobs must be less than the length of the XF job, or the extra time will accumulate for each antenna, losing much of the time benefits. (As long as it is less, the time spent on just pueoSim should be invariant under an increase in population)
3. The number of core hours spent on pueoSim jobs will be roughly the same for the same number of neutrinos as each job is shorter (except for a small contribution of the job overhead taking a higher percentage of total time for shorter jobs)
4. Initially I had thought that maybe queuing pueoSim jobs while running the XF jobs could slow down the queue for the XF jobs. So, I made the loop wait to submit the batch pueoSim jobs until we had space for all of them to be active with the ~250 max jobs per user.
Additionally, while observing my many tests, I didn't notice any correlation between the number of CPU peuoSim jobs in the queue and the number of GPU XF jobs out of the queue.
5. Branch I'm developing this on is here
6. The total time for the test generation was 15.4 hours, which is slightly longer than the ~14 hours from the 2023_05_08 run. However, this test also used double the population size, larger values for the range of antenna heights (on average about 3x taller), and 20x more neutrinos simulated per antenna. So, the actual speed is better than it first looks.
|
|
|
198
|
Fri Feb 24 10:05:38 2023 |
Alex M | Paper Symmetric Run for Reviewer | We began a new run for a symmetric bicone antenna. The purpose of this is to satisfy the most recent comments from the reviewer on the paper. We are using the same GA as for the paper with the same parameters. I will update this post soon with more details about this run. There is a run_details file attached, but please wait for me to update this post with more specifics as the version of the GA being used is less connected to the details presented in that file.
The data can be found here: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_02_20_Symmetric_Run . |
| Attachment 1: run_details_2023_02_20_Symmetric_Run.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_02_20_Symmetric_Run' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=0 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=0 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=0 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=0 ## If 1, evolve curved sides. If 0, sides are straight
A=0 ## If 1, A is asymmetric
B=0 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=1 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=25 ## Probability of mutation (divided by 100)
SIGMA=6 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=8 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=0 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
Draft
|
Mon Apr 24 13:09:36 2023 |
Dylan Wells | PUEO Plots Status | Plots we want to have for PUEO:
FScorePlot2D
Fitness_Scores_RG
VariablePlot (PUEO equivalent of LRT plot)
Veff_Plot (currently the veff is equivalent to the fitness score for PUEO)
Veffectives_RG
Rainbow_Plot
Violin Plot
avg_freq
gain_vs_freq
polar_plotter
physics of results
Next steps:
Get the Error values out of the pueoSim root output (talking to Will)
Use output uan files from the new two-run XF simulation to test the frequency plots. (/fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_04_21_Test_6)
Implement the physics of results plots into the loop (Talk to Bryan / Dennis)
Get the average Veff/Fitness score from the default gain pattern in pueoSim to compare our results to. |
| Attachment 1: Rainbow_Plot.png
| 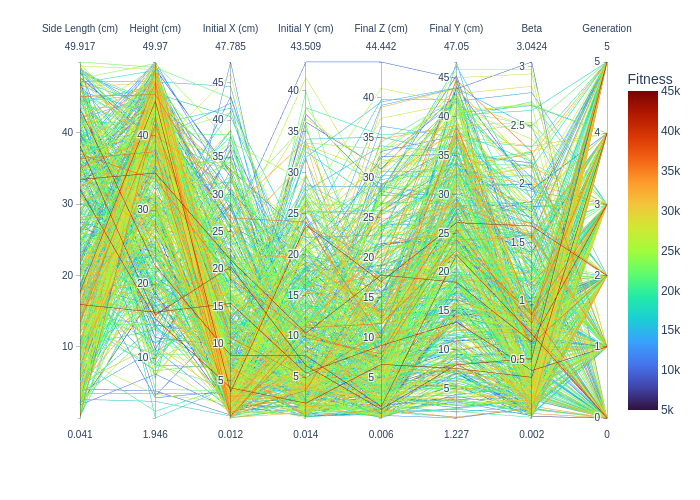 |
| Attachment 2: VariablePlot.png
| 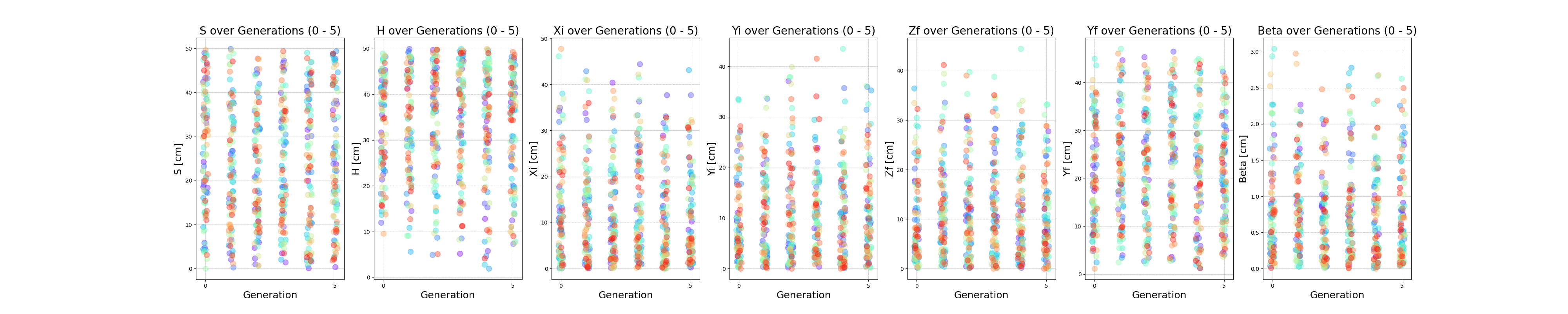 |
| Attachment 3: Fitness_Scores_RG.png
|  |
| Attachment 4: FScorePlot2D.png
|  |
| Attachment 5: Veff_plot.png
|  |
| Attachment 6: Veffectives_RG.png
|  |
| Attachment 7: ViolinPlot.png
| 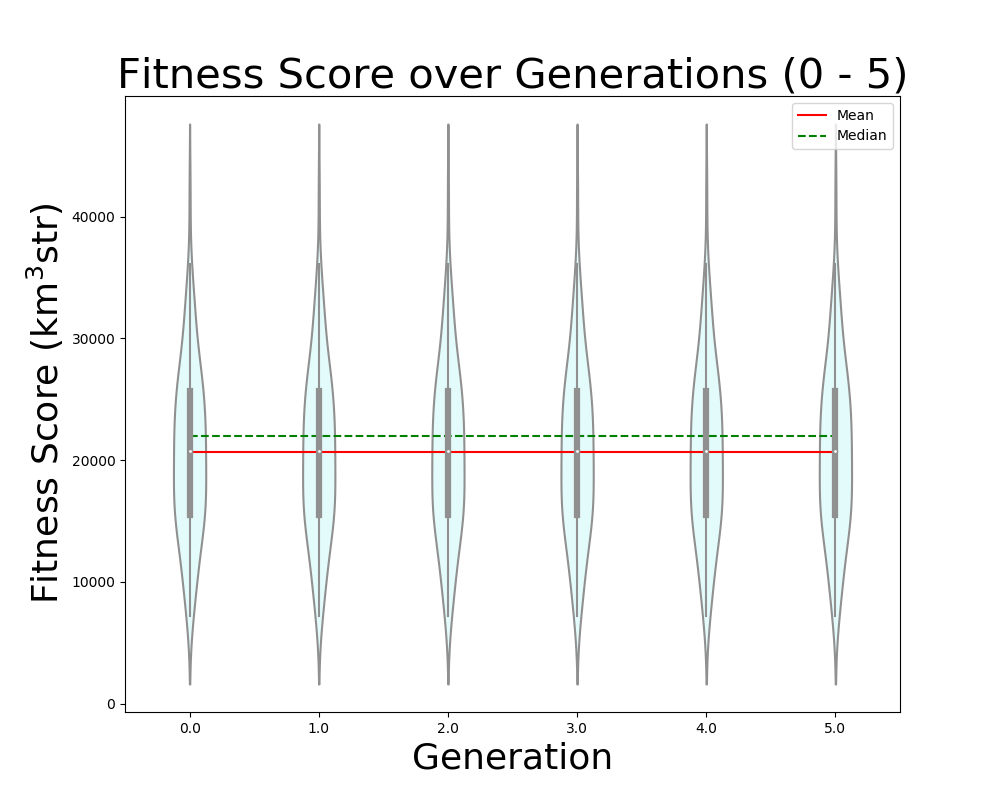 |
|
|
225
|
Wed Jun 14 14:38:22 2023 |
Alex M | PUEO Antenna Dimensions | Here are my recommended starting genes for the initial generation of the upcoming run (confirmed that these will produce a working antenna in XF). We want to use the lowest values we can because we expect that a larger (namely, taller) antenna will perform better:
S, H, x0, y0, yf, zf, beta
7.50000,50.0000,1.0000,1.00000,1.00000,50.0000,0.2
- S: Half the side length of the bottom of the antenna
- H: height of the antenna
- x0: distance of the bottom of the ridge from origin
- y0: width of ridge at the bottom
- yf: width of ridge at top
- zf: final height of the ridge (cannot exceed H)
- beta: a curvature parameter for the ridges
|
|
|
3
|
Wed Feb 6 17:30:51 2019 |
Julie Rolla | Our Github, Dropbox, & Slack links | Slack: https://gpantennas.slack.com/messages
elog:http://radiorm.physics.ohio-state.edu/elog/GENETIS/
Github link: https://github.com/mclowdus/BiconeEvolution
DropBox link: https://www.dropbox.com/home/GP_Antennas
Old GitHub for the evolution of the dipole: https://github.com/hchasan/XF_Scripts |
|
|
185
|
Thu Nov 10 10:42:20 2022 |
Ryan Debolt | Optimizations | This is the plot for the run type that had the best performance in the most recent optimization run that was completed. The most optimized runs from these collections are decided by which run types achieve a chi-squared value of 0.1 in the fewest generations (correlating to about a 0.9 fitness score). However, the genetic algorithm does not use Chi-squared directly, instead trying to maximize the fitness score (which uses a chi-squared in the denominator) that simulates the error we see in arasim. This fitness score is usually maximized at 1.0 but the simulated error can push the score slightly beyond this. This test was conducted with a simulated error of 0.1. This optimization holds the selection methods steady at 2 tournament 2 roulette and 6 rank selected individuals per 10 selection events. The changing parameter values are in the amounts of Reproduction, Crossover, Immigration, and mutation (rate and width) that the GA performs each generation. Each generation uses 100 individuals. In the graph below, the dotted lines are the minimum Chi^2 values at each generation, solid lines are the maximum fitness score. Each color represents a different run using the same parameters and dotted and solid lines sharing the same color represent the same run. The prevailing trend in the tests leading up to this test has been that the most optimized runs have low amounts of reproduction, high amounts of crossover, and they have been pushing the boundaries of the mutation rate and gaussian width of those mutations and this run follows that trend. The ranges of values we tested over were 0-12R, 88-100C, 15-25M_Rate, and 3-7 sigma, up from the previous test. This run took an average of 16.8 +/- 11.9 generations to reach the 0.1 chi-squared benchmark, the next closest run took 20.3 +/- 10.9 generations to reach the same point, while the most optimized run from an earlier test came in as the 10th best run taking 21.9 +/- 14.4 generations. The graph shows the behavior of this by showing large improvements in early generations that tend to quickly plateau around the maximum value before oscillating around the peak it reaches. Oscillations could be caused by either small mutations around our answer and displacement from the simulated error. The Chi-squared values show a much smoother behavior with smaller oscillations at the end of the run, which helps demonstrate the impact of the error on the behavior. |
| Attachment 1: Plot0_96_25_6.png
| 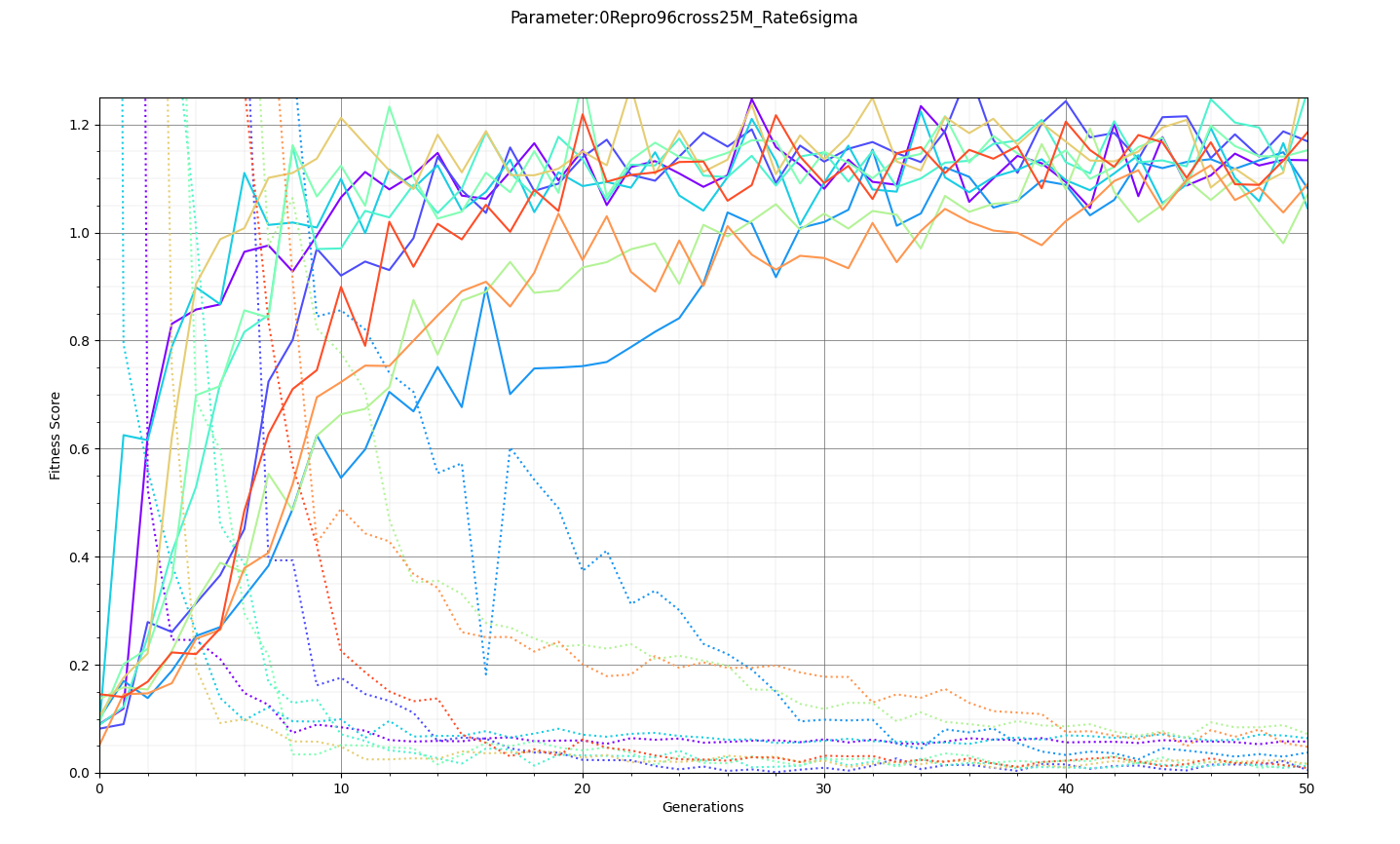 |
|
|
227
|
Wed Jun 28 15:49:44 2023 |
Ryan Debolt | Optimization Runs 6/12/2023 | All data and plots discussed in this post are taken from this spreadsheet:
https://docs.google.com/spreadsheets/d/1BbpD81mugWQf10ozGDLI60takAn3tlvrq8ksT10yV5I/edit?usp=sharing
Run Details:
This optimization run was done to do a course search for the optimal set of parameters to run the GA with. This run used a fixed 100-individual population with a simulated error of 0.25 over 50 generations, to best replicate the current PUEO-loop environment. The selection methods have also been held fixed in this run. This run was tested using both the ARA and PUEO design to see if there were similarities in the best run type. This run encompassed 210 different run combinations that searched the following parameter space:
- 0-16 Reproduction, step sizes of 4
- 72-96 Crossover, step sizes of 4
- 4-16 Mutation, step sizes of 4
- 5-15 Sigma, step sizes of 5
Results:
Each run combination was run 10 times and we tracked the average number of generations it took for the distance metric to reach 0.05, which corresponds to a 0.95 true fitness score. The best runs for each design were as followed:
- ARA
- Parameters
- 12 Reproduction
- 72 Crossover
- 4 Mutation
- 5 Sigma
- Average gens to benchmark
- Standard Deviation
- PUEO
- Parameters
- 0 Reproduction
- 96 Crossover
- 4 Mutation
- 15 Sigma
- Average gens to benchmark
- Standard Deviation
We can see that these run types do not appear to have any strong correlations. In fact, looking at our complete data set, there does not appear to be a strong trend around any of the best runs. This issue became more prominent when we ran these best two run types for 100 tests rather than 10. When we did this, they returned averages of 44.4 generations for PUEO and 46.9 generations for ARA. For comparison, the average number of generations across all run types was 43.1 generations for PUEO and 42.8 generations for ARA. This seems to suggest that all of these run types are inherently inconsistent and regress to the mean given enough tests. Therefore, I do not believe we can draw any conclusions from this run.
Moving Forward:
Our ongoing hypothesis is that the size of the error could be causing inconsistency in how quickly our population can grow over time. Therefore, our next step is to repeat this experiment, but with zero error included, and see if we achieve results that show consistent behavior. If we can find more consistency in runs with no error, then we can more deeply explore the effects that error can have on GA growth. If there continues to be a lack of consistency, then we can look to other factors, such as population size, and try to find the root of the inconsistency.
|
|
|
242
|
Sun Oct 20 13:11:09 2024 |
Dylan Wells | OSU Physics Scholarship Opportunities | Here are some scholarships available to physics majors I've been lucky enough to receive throughout undergrad that allowed me to work on this project unpaid without needing supplemental income from a separate job or loans.
1. OSU Arts and Sciences Merit Scholarship Pooled Application
- 500 word personal statement
- 1 letter of recommendation from a faculty member (Amy wrote mine)
- Varying amounts awarded (I got the $3,300 David and Velva Zarley Scholarship)
- Can be used for any education related expensed (tution, rent, food, ...)
2. James L. Smith Scholarship for Physics Majors
- 3 short essays (~200 words)
- Probably varying amounts (I got $4,000)
- Can be used for any education related expensed (tution, rent, food, ...)
3. Physics Class Awards
- No application, chosen by department (just get good grades, honors might help. I didn't really talk to any of my professors or go to office hours, so milage might vary there)
- Freshman: $50, Sophomore: $250, Junior: $500, Senior: $?
- Can be used for any education related expensed (tution, rent, food, ...)
4. OSU Scholarship Universe
- A couple ~500 word essay
- Very few awarded, I've applied twice: $1,000 the first time and completely ghosted the second
- Can be used for any education related expensed (tution, rent, food, ...)
5. Licking County Foundation (If anyone is from licking county, I HIGHLY RECOMMEND APPLYNG TO THIS. I think there are similar ones for other counties)
- ~500 word essay
- Varying awards (I've applied 3 times and gotten $2,375, $5,000, and $7,500)
- Can be used for any education related expensed (tution, rent, food, ...)
Here is a link to a google drive with all my winning scholarship essays:
https://drive.google.com/drive/folders/1qJKpBf4by9wlReU5l_XjxjVsDIfXR4Jc
|
|
|
238
|
Mon Aug 21 15:47:13 2023 |
Amy | OSC license agreement to be able to use XF | Attached is the license agreement that each person should sign to be able to use XF on OSC. You can sign it, send it to Amy, and she will return it to OSC with her signature on it. |
| Attachment 1: User_Software_Agreement-1.pdf
|
| Attachment 2: xfdtd.pdf
|
|
|
41
|
Wed Mar 18 15:01:10 2020 |
Julie Rolla | ONBOARDING: Tutorial Projects, Reading, Setup, etc. for New Students | For each new student joining the group, we ask them to onboard by completing each of the following items in the order given:
(1) Go to osc.edu and request an account
(2) Complete the following tutorials and reading. Note that the order of these does not matter, however, I would suggest "how to use Git" comes last, as it is the lease important for you to start. Finally, I advise you work on a tutorial concurrently with reading the pages assigned below from my Dissertation.
- OSC tutorials
- Because this project uses a lot of computation power, we run on the Ohio Super Computing Center's clusters. Please utilize these tutorials to learn more.
- Read Julie's Dissertation pages 1-9, 12-20 (starting at section 1.5.2), 28-37 (starting at section 2.3)
- Feel free to read chapter 3, as it has the history of this project and goes into details of our most up-to-date endeavors. This is optional and not mandatory.
- Complete the introduction to Bash project
- Complete the introduction to Python Project
- How to use Git (optional)
- Make sure to get an XFdtd license on OSC
(3) Finally, review the full manual and complete the bullets below. The full manual is Appendix A of Julie's Dissertation.
- Appendix A of my dissertation is our manual and acts as the holy grail of our research. Anything you need can be found here (from where our info exists on the web, to how to run the code).
- Once you have read this through entirely, we advise you log on to OSC and explore the code for PAEA in PAS1960. Once you have done so, complete this homework by making a copy of this google doc and filling it out on your own.
Once these are completed, the students are ready to start helping. This is all of the coding information and basic background information on evolutionary algorithms students must know to be proficient in assisting.
Other useful materials we have may include:
- Our old GENETIS manual
- This is somewhat out of date; however, it does have information on the general working on the loop.
- Julie's candidacy paper
- Complete the writing a genetic algorithm in C++ project
- This is still a work in progress! I'll complete it shortly!
- Read chapter 3 of this book (more if you have time) by Eiben and Smith, "Introduction to Evolutionary Computation" for a quick introduction. The book should be available electronically from a university library. Let us know if you can't find it there.
- Get to know XFdtd
- XFdtd manual is also attached; however, the link above (in 5) is more useful for coding a .xmacro.
Important notes:
MAC USERS: Using a GUI when connecting to OSC via SSH from your terminal.
You will need to set up X11 forwarding. Otherwise, when you go to plot via SSH, they will not display (they don't forward from OSC to your machine). If you are unable to run GUI's on OSC through your computer's terminal, follow these instructions:
- In your own system's terminal, type: sudo vi /etc/ssh/sshd_config
- Search for the following line: X11Forwarding no
- Change it to: X11 Forwarding yes. Save the file and exit.
- In your own system's terminal, type: sudo systemctl restart sshd
- For a Mac, download and install XQuartz.
- Connect to OSC using ssh -XY username@hostname.
- You can test that this works by typing 'xeye' or 'xclock' in the terminal when logged into OSC.
Now you should be able to use OSC's GUIs.
PC USERS:
As a PC user, there are a few things you need to do:
(1) Enable the Windows Subsystem for Linux on your device (https://developerinsider.co/stepwise-guide-to-enable-windows-10-subsystem-for-linux/#:~:text=To%20enable%20the%20Windows%20Subsystem,list%20here%20and%20click%20OK.)
(2) Get Bash on Ubuntu installed on your machine. You are going to need to have Bash on Ubuntu installed prior to starting these trainings; otherwise Bash commands will not be recognized.
(3) Get X11 forwarding working. Otherwise, when you go to plot via SSH, they will not display (they don't forward from OSC to your machine).
- Follow these steps here.
- Change your config file. To do so, in terminal (with Bash on Ubuntu open)
- type: sudo vi /etc/ssh/sshd_config
- Then change '#X11Forwarding no' to '#X11Forwarding yes' and save it.
- Make sure Putty and Xming are running every time before you ssh in. Use Xlaunch application to run Xming and press yes to everyrthing.
- Xming should now be running as a background task.
- Open PuTTY and open the pre-saved like you saved in PuTTY. (This will open a Bash terminal that asks for your OSC login password.)
|
| Attachment 1: 237799239-XFdtd-Reference-Manual-pdf.pdf
|
|
|
9
|
Wed Feb 27 16:29:35 2019 |
Julie Rolla | New To-Do List | Here are the following things we need to work on:
- Paper (Julie and Suren)
- Get loop working-- specifically XF (Cade)
- Test GA (Suren)
- Gain pattern plots -- Make them 3D (Evelyn) & put on GitHub
- Insert LR plot & Gain plot to loop (?)
- Edit bash script
- Update Github
- AraSim plots (Julie and Max) -- see #2 and #3 here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/7 for details.
- Update manual with things from Suren's last post (?)
|
|
|
131
|
Fri Mar 19 17:39:47 2021 |
Alex M | New Run | We began a new run using the same parameters as the previous one (see the last ELOG entry). The previous one ran for 8 generations. The difference between this run and the last one is that we have added in the polarity fix for AraSim Brian and Jorge gave us (copying the correct Report.cc and Report.h files into our AraSim version). Here are the run parameters:
Run Paramaters:
50 individuals
80/20 Roulette/Tournament
6% Reproduction, 72% Crossover, 22% Mutation
Run for 10 generations, then implement Jorge and Brian's fix for polarization in AraSim for the next run |
|
|
Draft
|
Thu Jun 30 13:04:48 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | This is a multigenerational tracing of our second-best individual's parents and children:
The second-best individual in this evolution was Bicone 22 from generation 40. This individual is, in fact, a fascinating case as we shall see. But to start the story of this individual we will go back to generation 38 in order to demonstrate some of the peculiarities.
In generation 38 there were two bicones of no renown, Bicone 11 and Bicone 17. Bicone 11 was a fairly average individual that was ranked 23rd with a fitness score of 4.72016. Its DNA was {*6.16084, ***79.663, 0.0015434, -0.107765} for side one , and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. One day, by chance it met Bicone 17, another average bicone ranked 24th with a score of 4.71877 and DNA {**2.32499, 79.663, -0.00224948, 0.192602} for side one, and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. These two individuals eventually became the parents of two antennas: Bicone 16 and Bicone 17 of generation 39.
Bicone 16 eventually grew to have been ranked 3rd in fitness score of 4.95323. It’s DNA ended up being a complete balance of its parents sharing side one with Bicone 38.11{2.32499, 79.663, -0.00224948, 0.192602} and side two with bicone 38.17 {0.308809, 39.6742, -0.0084247, 0.40629}. Bicone 16 was an individual with high aspirations and hoped to be reproduced. But alas, it was not meant to be. But bicone 16 came upon some amazing luck, it was selected with itself for crossover. This meant that bicone 16 was able to provide two identical surrogates to survive into the next generation. This is where this Bicone fulfilled its full potential.
The twin bicones were named Bicone 22 and Bicone 23 in generation 40. Being clones, they shared all their DNA with Bicone 39.16. However, due to some circumstances, they had slightly different fitness scores. Bicone 23 managed a very respectable 5.0117 fitness score and was ranked 2nd in the generation. But not to be outdone Bicone 22 managed to score a 5.17014 and ended up being the second-best performing individual of all time. Being so successful, the two bicones ended up producing 8 children between the two groups.
Bicone 23 was the first to crossover and had 2 children with its partner. These were Bicones 4 and 5. Bicone 4 was ranked 39th with a fitness score of 4.60251 and still shared the DNA of its second side with its grandparent Bicone 38.17 as well as most of its first side with Bicone 38.11 {2.32499, 79.663, -0.00213879, 0.192602} {0.308809, 39.9608, -0.0084247, 0.40629}. Bicone 5 on the other hand, was ranked 14th with a fitness score of 4.80535 and it still shared a lot of DNA with its grandparents {6.16084,53.0851,-0.00224948,0.0534469} {0.966617,39.6742,-0.0084247,0.40629}.
Bicone 22 had 6 children of its own with various other Bicones. These were; Bicone 8, ranked 11th with a fitness Score of 4.81784 and DNA {2.32499, 75.9855, -0.00224948, 0.192602} {0.966617, 39.6742, -0.00320023, 0.213833}; Bicone 9, ranked 7th with a fitness score of 4.88966 and DNA {6.10508, 79.663, -0.000594616, 0.0351901} {0.308809, 42.4246, -0.0084247, 0.40629}; Bicone 12, ranked 12th with a fitness score of 4.81705 and DNA {6.42695, 75.9855, 0.0015434, -0.107765} {0.308809, 39.6742, -0.00320023, 0.213833}; Bicone 13, ranked 2nd with a fitness score of 5.0344 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.966617, 42.4246, -0.0084247, 0.40629}; Bicone 34 ranked 3rd with a fitness score of 4.99864 and DNA {0.66148, 73.5522, -0.000594616, 0.00582814} {0.966617, 39.6742, -0.0084247, 0.40629}; and finally Bicone 35, ranked 22nd with fitness score 4.74955 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.308809, 42.4246, -0.0084247, 0.40629}
Bellow, I have attached the rainbow plot with the parameters occupied by individual 4 in gen 41 which was again ranked 39th in that generation. From this, we can see that while in its own generation it was a poor performer, overall it was upper middle of the pack. However, because of the density of other better performing antennas in this region, it is hard to distinguish which genes in this antenna are contributing the most to the drop in fitness score compared to its siblings and parents.
*Gene originating from Bicone 38.11
**Gene originating from Bicone 38.17
***Gene originating from Bicone 38.11 and 38.17 that is shared between the two. |
| Attachment 1: rainbow_plot_quadratic_rank_test_720_(drawn).png
| .png) |
|
|
Draft
|
Fri Jul 8 13:34:08 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | |
|
|
173
|
Fri Jul 8 13:35:19 2022 |
Ryan Debolt | Multigenerational Narrative draft 2 | The second-best individual in this evolution was Bicone 22 from generation 40. This individual is, in fact, a fascinating case as we shall see. But to start the story of this individual we will go back to generation 38 in order to demonstrate some of the peculiarities.
In generation 38 there were two bicones of no renown, Bicone 11 and Bicone 17. Bicone 11 was a fairly average individual that was ranked 23rd with a fitness score of 4.72016. Its DNA was {*6.16084, ***79.663, 0.0015434, -0.107765} for side one , and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. One day, by chance it met Bicone 17, another average bicone ranked 24th with a score of 4.71877 and DNA {**2.32499, 79.663, -0.00224948, 0.192602} for side one, and {0.308809, 39.6742, -0.0084247, 0.40629} for side two. These two individuals eventually became the parents of two antennas: Bicone 16 and Bicone 17 of generation 39.
Bicone 16 eventually grew to have been ranked 3rd in fitness score of 4.95323. It’s DNA ended up being a complete balance of its parents sharing side one with Bicone 38.11{2.32499, 79.663, -0.00224948, 0.192602} and side two with bicone 38.17 {0.308809, 39.6742, -0.0084247, 0.40629}. Bicone 16 was an individual with high aspirations and hoped to be reproduced. But alas, it was not meant to be. But bicone 16 came upon some amazing luck, it was selected with itself for crossover. This meant that bicone 16 was able to provide two identical surrogates to survive into the next generation. This is where this Bicone fulfilled its full potential.
The twin bicones were named Bicone 22 and Bicone 23 in generation 40. Being clones, they shared all their DNA with Bicone 39.16. However, due to some circumstances, they had slightly different fitness scores. Bicone 23 managed a very respectable 5.0117 fitness score and was ranked 2nd in the generation. But not to be outdone Bicone 22 managed to score a 5.17014 and ended up being the second-best performing individual of all time. Being so successful, the two bicones ended up producing 8 children between the two groups.
Bicone 23 was the first to crossover and had 2 children with its partner Bicone 25 (4.64409). These were Bicones 4 and 5. Bicone 4 was ranked 39th with a fitness score of 4.60251 and still shared the DNA of its second side with its grandparent Bicone 38.17 as well as most of its first side with Bicone 38.11 {2.32499, 79.663, -0.00213879, 0.192602} {0.308809, 39.9608, -0.0084247, 0.40629}. Bicone 5 on the other hand, was ranked 14th with a fitness score of 4.80535 and it still shared a lot of DNA with its grandparents {6.16084,53.0851,-0.00224948,0.0534469} {0.966617,39.6742,-0.0084247,0.40629}.
Bicone 22 had 6 children of its own with various other Bicones. These were; With Bicone 8.40 (4.82423) Bicone 8, ranked 11th with a fitness Score of 4.81784 and DNA {2.32499, 75.9855, -0.00224948, 0.192602} {0.966617, 39.6742, -0.00320023, 0.213833}; Bicone 9, ranked 7th with a fitness score of 4.88966 and DNA {6.10508, 79.663, -0.000594616, 0.0351901} {0.308809, 42.4246, -0.0084247, 0.40629}; With Biocone 16.40 (5.01137) Bicone 12, ranked 12th with a fitness score of 4.81705 and DNA {6.42695, 75.9855, 0.0015434, -0.107765} {0.308809, 39.6742, -0.00320023, 0.213833}; Bicone 13, ranked 2nd with a fitness score of 5.0344 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.966617, 42.4246, -0.0084247, 0.40629}; With Bicone 4.40 (4.66578) Bicone 34 ranked 3rd with a fitness score of 4.99864 and DNA {0.66148, 73.5522, -0.000594616, 0.00582814} {0.966617, 39.6742, -0.0084247, 0.40629}; and finally Bicone 35, ranked 22nd with fitness score 4.74955 and DNA {2.32499, 79.663, -0.00224948, 0.192602} {0.308809, 42.4246, -0.0084247, 0.40629}
Upon inspection of the children in generation 41, you will see that none of the children surpassed their parent individual with the highest fitness score. They do however tend to have fitness scores around or above their second parent's score. Therefore, it is likely that the children's lower fitness score is due to a mismatch of genes. If we look at the rainbow plots provided, we can see that the genes of the worst-performing child are not dissimilar to those of individuals that scored higher than 5. This would imply that the fitness score found could be due to variations in Arasim similar to those seen from individuals 16.39 to 22/23.40 despite them being identical. Though it also could be possible that the graphs do not have a fine enough display to be able to clearly tell some of the genes apart.
*Gene originating from Bicone 38.11
**Gene originating from Bicone 38.17
***Gene originating from Bicone 38.11 and 38.17 that is shared between the two.
Amy: We want to nail things down in places where we still have hypotheses for what could have happened. For example, "This would imply that the fitness score found could be due to variations in Arasim similar to those seen from individuals 16.39 to 22/23.40 despite them being identical. Though it also could be possible that the graphs do not have a fine enough display to be able to clearly tell some of the genes apart." These two hypotheses can easily be distinguished by running with higher statistics and looking at the numbers rather than relying on the resolution of the display.
Amy: Also, at each stage, I was looking for the random numbers that were generated that led to the selection of the fractions of the population that were used for each type of selection and operator.
|
| Attachment 1: Rainbow_Plot_Quadratic_Rank_Test_5.png
|  |
| Attachment 2: Rainbow_Plot_Quadratic_Rank_Test_4.5.png
|  |
| Attachment 3: Rainbow_Plot_Quadratic_Rank_Test_4.png
| 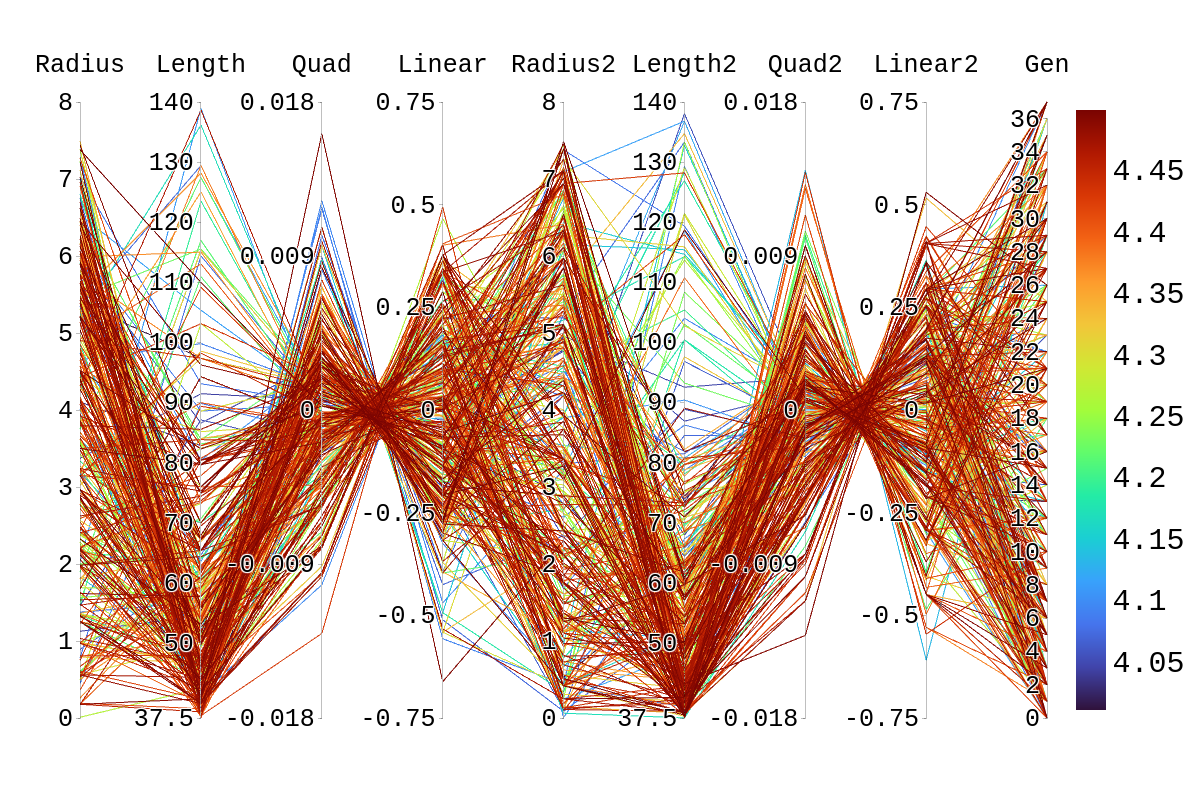 |
| Attachment 4: Rainbow_Plot_Quadratic_Rank_Test_3.5.png
| 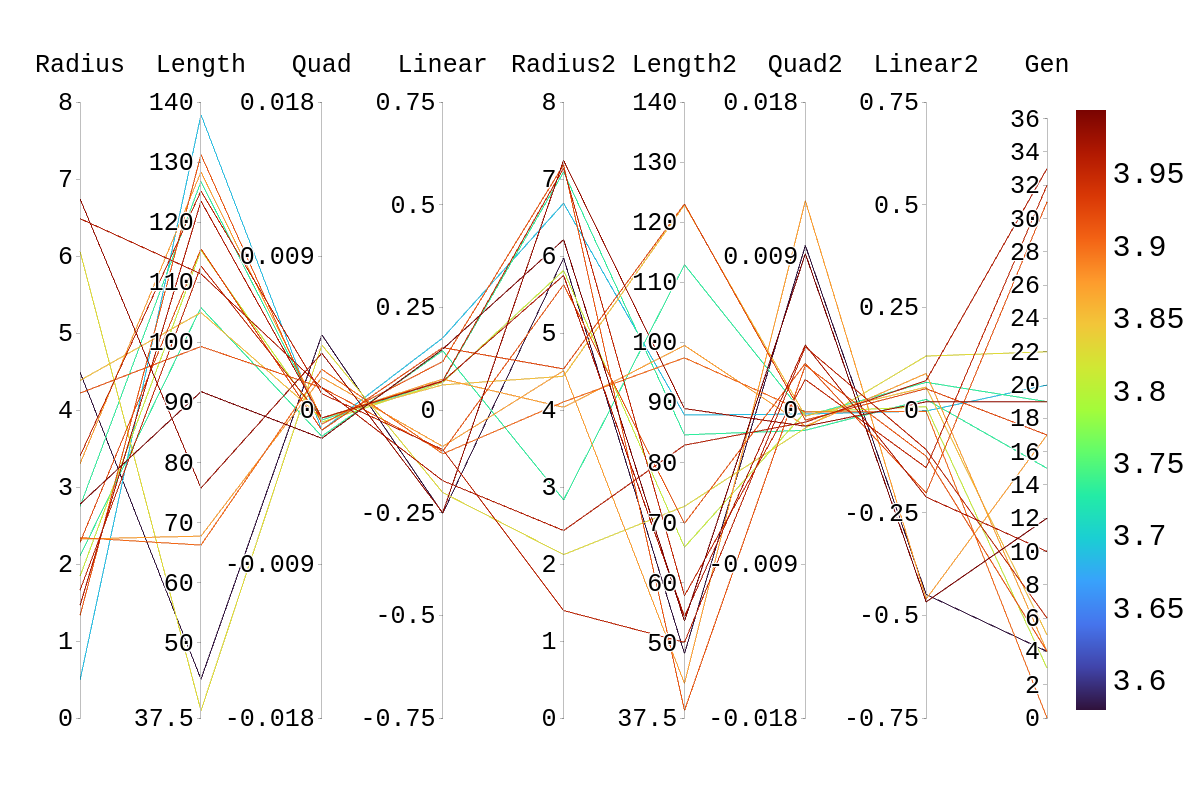 |
|
|
166
|
Tue Jun 21 13:20:41 2022 |
Ryan Debolt | Multigenerational Narrative draft | The story of individual 8 from generation 18. (draft)
Once, there was a curved bicone named individual 8 who was from generation 18. In many ways, it was similar to many other bicones but in fact, this individual was the best bicone that ever lived with a fitness score of 5.22097. It achieved this by having the following parameters; an inner radius of 6.31483, and 3.97024; lengths of 77.6017 and 40.9244; quadratic coefficients of, 0.00260615 and, -0.0103313; and finally, a linear coefficient of -0.197494 and
0.36119. Unfortunately, individual 8’s lineage ended with it as it never met that special someone (crossover) and didn't survive into the next generation (reproduction) despite having a 77.7% of doing at least one of these.
This bicone had a sibling with a fitness Score of 4.65861 and had the following parameters: 6.05614,79.663,-0.000353038,0.0331969 (side 1)
1.92878,37.9849,-0.00213265,0.156724 (side 2).
Unfortunately, this bicone also suffered the same fate as its sibling and failed to leave a lasting mark on this hypothetical world.
These two individuals had parents from generation 17 whose name’s were individuals 39 and individual 8. After having individuals 8 and 9 in generation 18, individuals 39 and 8 from generation had a nasty divorce (possibly leading to 8 and 9’s aversion to marriage) and remarried. In these marriages, they both produced two more children that would be our previous antenna’s step-siblings.
Antenna 8 from gen 17 married antenna 46 to produce antenna 20 with a fitness score of 4.57122, and antenna 21 with a fitness score of 4.71056. Individual 20 shared the same genes for the second set of linear and quadratic coefficients as its more successful step-sibling individual 8. 21 had no similarities to individual 8 but had 4 children in generation 19.
Antenna 39 from gen 17 married antenna 5 to produce antenna 28 with a fitness score of 4.71808 and antenna 29 with a fitness score of 4.90119. Antenna 28 shared had an identical side 1 to individual 8 and also shared the same inner radius and length on the second side, and had 2 children in generation 19. Individual 29 had no similarities to individual 8 but really got around and had 8 children with various partners in generation 19. |
|
|
120
|
Mon Nov 23 18:02:40 2020 |
Ryan Debolt | Monday Updates |
| Alex M |
Kept working with Amy, Alex P, Julie, and Ben on the AraSim fix. We fixed our issue from last week but have a new one in stage 2. It looks like the issue has to do with resetting the values for V_forfft right before stage 2 (around line 963). Check here for the current version of Report.cc: /users/PAS0654/pattonalexo/EFieldProject/11_23_20 |
| Ryan |
Fixed the issue with the segmentation faults in the GA (simple fix one line changed) and worked with Kai to start creating a spreadsheet of results. Results seem to suggest that 2 tournament to 8 roulette seems to be the ideal selection ratio and the initial test seem to suggest a small amount of reproduction is ideal but more testing is needed to confirm this. |
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
122
|
Mon Nov 30 17:00:31 2020 |
Ethan Fahimi | Monday Updates |
| Alex M |
Kicked the loop back up. Helped Ethan and Parker with their projects during the working meeting. |
| Ryan |
Changed the tournament/roulette ratio, reproduction_no, and crossover_no to be read in variables in the GA to increase the quality of life when running the algorithm and to prep the code for some testing. Format for how to call the code is written at the top of the cpp program file. |
| Ethan F |
Worked with Alex M on further fixing AREA. The first generation works now, we are making minor fixes to get subsequent generations up and running. |
| |
|
| |
|
| |
|
| |
|
|
|
|
215
|
Mon May 1 05:45:02 2023 |
Alex M | Midscale PUEO Run | I've started a run of the PUEO loop to get some data for evolving the antenna width and height. We are running with 24 individuals, each with 400k neutrinos at an energy exponent of 19. Here is the output directory: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_05_01_Test_5
It's very important to note that the cross-pol data being used still comes from the data already stored in pueosim. This is because our extremely low cross polarizations were causing us to have no effective volumes. There's likely some issue in what values from XF we are choosing as our crosspols. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_05_01_Test_5' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=10 ## number of generations (after initial) to run through
NPOP=24 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=40000 ## Number of Neutrinos Thrown in AraSim
exp=19 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=6 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
PUEO=1 ## IF 1, we evolve the PUEO quad-ridged horn antenna, if 0, we evolve the Bicone
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=6 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=10 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=2 ## Percent of individuals selected through tournament (divided by 10)
RANK=6 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
|
|
|
161
|
Tue Jun 7 14:14:21 2022 |
Dylan Wells | Matching Circuits Slides | Slides contatining my notes on matching circuits.
https://docs.google.com/presentation/d/1x25nhiqaW7LvPZ1pNZ5O4ZzsWZbtgqxBQ5haB9uWgQY/edit?usp=sharing |
|
|
191
|
Mon Feb 6 10:23:53 2023 |
Dylan Wells | Matching Circuit Parts | Attached is a spreadsheet with the information on the parts we need for the N=14 matching circuit board.
https://docs.google.com/spreadsheets/d/1x8dX3tNE-WSHjH_slj_EH4XsHcAnCVC05XiRBFPtIUc/edit#gid=0 |
|
|
137
|
Wed Sep 8 16:36:33 2021 |
Alex M | MODE Workshop Presentation | We presented at a workshop put on by the MODE collaboration. MODE is a collaboration dedicated to applying Automatic Differentiation to detector design. Here's the website: https://mode-collaboration.github.io/#:~:text=MODE%20(for%20Machine%2Dlearning%20Optimized,in%20space%2C%20and%20in%20nuclear
|
| Attachment 1: GENETIS_MODE_Presentation.pptx
|
|
|
142
|
Fri Feb 4 16:50:09 2022 |
Ryan Debolt | Loop Run |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- To test rank selection in main loop
- What is different about this run from the last?
- Rank Slection is being used.
- Parents.csv introduced.
- Elite is being turned off.
- Symmetric, asymmetric, linear, nonlinear (what order):
- Number of individuals (NPOP):
- Number of neutrinos thrown in AraSim (NNT):
- Operatiors used (% of each):
- 06% Reproduction
- 72% Crossver
- 22% Immigration
- 1% M_rate (unused)
- 5% sigma (unused)
- Selection methods used (% of each):
- 0% Elite
- 0% Reproduction
- 10% Tournament
- 90% Rank
- Are we using the database?
Directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_02_04_Rank |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_02_04_Rank' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=0 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=1 ## Percent of individuals selected through tournament (divided by 10)
RANK=9 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
128
|
Fri Jan 29 15:59:21 2021 |
Alex M | Loop Demonstration Video | Here's the video I made a few months ago demonstrating how to run the loop. There might be some changes that have been made since then, but they'll generally be minor so it should still be representative of what running the loop looks like. It might need to be shared with you through the Microsoft drive service OSU gives us in order to view it, so either message me on Slack or email me at machtay.1@osu.edu and I'll share the link through there.
https://drive.google.com/file/d/1yL_eH_w2hXNt7J7YOwS5xl3ZHwGyl5JR/view |
|
|
5
|
Thu Feb 21 14:07:14 2019 |
Julie Rolla | Logging into Nutau via XRDP | As mentioned in the manual, using ssh to login to Nutau causes delays; the XFdtd GUI cannot be surpress and must be forwarded via X11 forwarding, and this is extremely slow. We are currently looking into using a newly suggested option, XRDP. Information will be added as this process continues. |
|
|
236
|
Wed Aug 2 12:49:57 2023 |
Dylan Wells | Line of Best-Fit Straightened Sides Antenna | Previously, we have tried to straighten the sides of the best-evolved curved antenna in elog 229. However, there were potential issues with how closely this line resembled the curve of the antenna.
So, I attempted to create another straightened sides antenna using a linear regression model to find the best fitting line for the curve to create an antenna.
I made a Python notebook to separate the equations for each curve into 1000 discrete points. Then I ran a linear regression model to fit a curve of the points 1 - n, and a second curve of the n - 1000 points, looping from n = 2 to n = 999.
These results are from the combined output with the least squared error compared to the original.
Pictured is a plot showing the two sides of the curved bicone in red and blue, with the best fitting lines for each in black as well as a model in XF.
Results:
The antenna has a fitness score of 3.80627 with an error of 0.0759725.
This is much lower than the 5.71 of the curved antenna and 5.11 of the other attempt at straightening.
For the next attempt, we could consider constraining the endpoints to be the same as the original antenna to conserve the radius, and/or adding an extra line to fit the curve.
I've also attached a picture of what my notebook found for fitting 3 lines to the curve (not modeled or tested).
Professor Chen recommended using 3 sides and constraining the outer radii of the cones to match the original curved design.
Path to linear regression notebook: /users/PAS1960/dylanwells1629/developing/notebook.ipynb
Path to XF project: /users/PAS1960/dylanwells1629/straightened.xf |
| Attachment 1: output.png
| 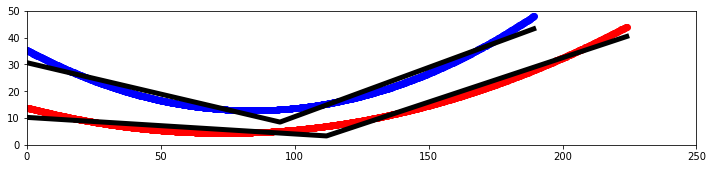 |
| Attachment 2: image_(3).png
| .png) |
| Attachment 3: output.png
| 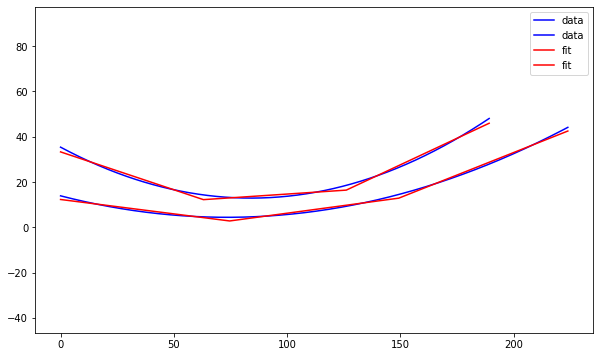 |
|
|
126
|
Tue Jan 19 19:00:30 2021 |
Alex M | Latest Run Details/Running on Slurm | OSC managed to figure out our issue with running XF from an interactive job on Slurm. Previously, we were losing our connection to x11 forwarding. The solution is to use sinteractive instead of srun to obtain an interactive job. Here's the syntax:
sinteractive -A PAS0654 -t <time> -N 1 -n 8 -p serial
The -p serial flag denotes the type of partition to request. It's important to specify this, as otherwise it will default to the debug node, which has a limit of 1 hour.
We're going to begin a new run. The title is Machtay_2021_1_15_NPOP50_Asym . It is using the latest version of the GA Ryan has been working on (Latest_GA_Asym.cpp). We're using 6% reproduciton, 72% crossover, and 22% mutation. We are using 80% roulette selection. This correlates to the three numbers passed into the GA being 3 36 8 (since we're using 50 individuals). |
|
|
179
|
Tue Aug 16 11:41:07 2022 |
Dylan Wells | Instructions for Running IceMC | Running IceMC:
Go into the directory ../anitaBuildTool/build/components/icemc/
And run the command
*May need to chmod -R 775 ../anitaBuildTool/comonents/icemc/ if you get a permissions error
inputFile:
Must be the full path to the file
Config files are found in ../anitaBuildTool/components/icemc
Ex: /users/PAS1960/dylanwells1629/anitaBuildTool/components/icemc/inputs.anita4.conf
Config files are found in ../anitaBuildTool/components/icemc
outputDirectory:
Will be made in ../anitaBuildTool/build/components/icemc/ by default, specify full path otherwise.
runNumber:
The run number.
numberOfNeutrinos:
The number of neutrinos generated in the simulation.
Can be found in inputs.conf
Default is 2,000,000.
#How many neutrinos to generate
triggerThreshold:
Threshold for each band for the trigger.
Default is 2.3
#thresholds for each band- this is only for the frequency domain voltage trigger. If using a different trigger scheme then keep these at the default values of 2.3 because the max among them is used for the chance in hell cuts
energyExponent:
The exponent of the energy for the neutrinos
Can be found in input.conf
Default is 1020
# Select energy (just enter the exponent) or (30) for baseline ES&S (1) for E^-1 (2) for E^-2 (3) for E^-3 (4) for E^-4 (5) for ES&S flux with cosmological constant (6) for neutrino GZK flux from Iron nuclei (16-22)not using spectrum but just for a single energy (101-114)use all kinds of theoretical flux models |
|
|
178
|
Wed Aug 10 22:38:20 2022 |
Dylan Wells | Instructions for Installing IceMC | Installing IceMC:
I: Getting anitaBuildTool
Clone the anitaBuildTool repository (https://github.com/anitaNeutrino/anitaBuildTool) to your user.
II: Get the Anita.sh file onto your user.
Either copy the file from /users/PAS1960/dylanwells1629/Anita.sh
or create a new file Anita.sh with the following code
# .bashrc
# Source global definitions
if [ -f /etc/bashrc ]; then
. /etc/bashrc
fi
#this modules were originally loading in both the env.sh, and bashrc_anita.sh files. This was redundant so it was added here, and removed from the others.
module load gnu/7.3.0
module load gnu
module load mvapich2
module load fftw3
#module load python/3.6-conda5.2
module load cmake
PATH=$PATH:$HOME/.local/bin:$home/bin
export PATH
export CC=`which gcc`
export CXX=`which g++`
export FFTWDIR=/fs/project/PAS0654/shared_software/fftw3/gnu/6.3/mvapich2/2.2/3.3.5
export ANITA_SOURCE_DIR=~/anitaBuildTool/
export ANITA_UTIL_INSTALL_DIR=~/anitaBuildTool/
export ICEMC_SRC_DIR=~/anitaBuildTool/components/icemc/
export ICEMC_BUILD_DIR=~/anitaBuildTool/build/components/icemc/
export DYLD_LIBRARY_PATH=${ICEMC_SRC_DIR}:${ICEMC_BUILD_DIR}:${DYLD_LIBRARY_PATH}
export ROOTSYS=/fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root
# User specific aliases and functions
#This env.sh is for running the BiconeEvolution GENETIS software. This should only be un-commented if you are running GENETIS software. When you do this, comment out env.sh.
#source ~/new_root_setup.sh
source /fs/project/PAS0654/shared_software/anita/owens_pitzer/build/root/bin/thisroot.sh
#source /cvmfs/ara.opensciencegrid.org/trunk/centos7/setup.sh
#module load python/3.6-conda5.2
#BiconeGENETIS directory shortcut SHARED
alias GE='cd ../../../fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/'
#emacs Alias
alias emacs='emacs -nw'
#root alias
alias root='root -l'
#Alias
alias l="ls"
alias python='/cvmfs/ara.opensciencegrid.org/trunk/centos7/misc_build/bin/python3.9'
Then source the file
Note: You will need access to PAS0654 for this step or you will get a permissions error.
III: Running the build tool.
Go into the anitaBuildTool directory
And run the building script
Note: There will be an error if you source files for running Ara in your .bashrc
Comment these out and restart your terminal before running the build. (remember to source Anita.sh before running the build tool. You could also source Anita.sh in your .bashrc)

Error if you source files for running Ara:
CMake Error at components/libRootFftwWrapper/cmake_install.cmake:238 (file):
file INSTALL cannot copy file
"/users/PAS1960/dylanwells1629/anitaBuildTool/components/libRootFftwWrapper/include/AnalyticSignal.h"
to
"/cvmfs/ara.opensciencegrid.org/v2.0.0/centos7/ara_build/include/AnalyticSignal.h":
Read-only file system.
Call Stack (most recent call first):
|
|
|
204
|
Mon Mar 20 16:31:09 2023 |
Alex M | Installing PUEOsim | Will sent me some info on how to install PUEOsim. I attached a markdown file he sent for how to install PUEOsim. He also sent the path to pre-compiled installation of PUEOsim. Here's the path to the pre-compiled installation: /users/PAS0654/wluszczak/PUEO_shared
Here is his full message, which details that you must also run set_env.sh before running PUEOsim:
a pre-compiled version is located at /users/PAS0654/wluszczak/PUEO_shared . Inside that directory, there's a set_env.sh script that you will need to source before running anything (hopefully this works, though there's a high probability you might run into permissions issues with the dependencies).
To run PUEOsim, use the following (see here for instructions: https://github.com/PUEOCollaboration/pueoSim):
./simulatePueo -i {inputFile} -o {outputDirectory} -r {runNumber} -n {numberOfNeutrinos} -e {energyExponent}
The outputs will be root files, but it should be possible (either modifying our own version directly or with a script) to print out the effective volume to a .txt to read.
|
| Attachment 1: CentOS_7_install.md
|
#pueoBuilder Installation Tutorial
This tutorial will guide you through the process of building the tools included in pueoBuilder from scratch, including the prerequisites and any environment variables that you will need to set. This sort of thing is always a bit of a nightmare process for me, so hopefully this guide can help you skip some of the frustration that I ran into. I did not have root acces on the system I was building on, so the instructions below are what I had to do to get things working with local installations. If you have root access, then things might be a bit easier. For reference I'm working on CentOS 7, other operating systems might have different problems that arise.
##Prerequisites
As far as I can tell, the prerequisites that need to be built first are:
-cmake 3.21.2 (I had problems with 3.11.4)
-gcc 11.1.0 (9.X will not work)
-fftw 3.3.9
-gsl 2.7.1 (for ROOT)
-ROOT 6.24.00
###CMake
You can download the source files for CMake here: https://cmake.org/download/. Untar the source files with:
tar -xzvf cmake-3.22.1.tar.gz
Compiling CMake is as easy as following the directions on the website: https://cmake.org/install/, but since we're doing a local build, we'll use the `configure` script instead of the listed `bootstrap` script. As an example, suppose that I downloaded the above tar file to `/scratch/wluszczak/cmake`:
mkdir install
cd cmake-3.22.1
./configure --prefix=/scratch/wluszczak/cmake/install
make
make install
You should additionally add this directory to your `$PATH` variable:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
To check to make sure that you are using the correct version of CMake, run:
cmake --version
and you should get:
cmake version 3.22.1
CMake suite maintained and supported by Kitware (kitware.com/cmake).
### gcc 11.1.0
Download the gcc source from github here: https://github.com/gcc-mirror/gcc/tags. I used the 11.1.0 release, though there is a more recent 11.2.0 release that I have not tried. Once you have downloaded the source files, untar the directory:
tar -xzvf gcc-releases-gcc-11.1.0.tar.gz
Then install the prerequisites for gcc:
cd gcc-releases-gcc-11.1.0
contrib/download_prerequisites
One of the guides I looked at also recommended installing flex separately, but I didn't seem to need to do this, and I'm not sure how you would go about it without root priviledges, though I imagine it's similar to the process for all the other packages here (download the source and then build by providing an installation prefix somewhere)
After you have installed the prerequisites, create a build directory:
cd ../
mkdir build
cd build
Then configure GCC for compilation like so:
../gcc-releases-gcc-11.1.0/configure -v --prefix=/home/wluszczak/gcc-11.1.0 --enable-checking=release --enable-languages=c,c++,fortran --disable-multilib --program-suffix=-11.1
I don't remember why I installed to my home directory instead of the /scratch/ directories used above. In principle the installation prefix can go wherever you have write access. Once things have configured, compile gcc with:
make -j $(nproc)
make install
Where `$(nproc)` is the number of processing threads you want to devote to compilation. More threads will run faster, but be more taxing on your computer. For reference, I used 8 threads and it took ~15 min to finish.
Once gcc is built, we need to set a few environment variables:
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
We also need to make sure cmake uses this compiler:
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/bin/gfortran-11.1
If your installation prefix in the configure command above was different, substitute that directory in place of `/home/wluszczak/gcc-11.1.0` for all the above export commands. To easily set these variables whenever you want to use gcc-11.1.0, you can stick these commands into a single shell script:
#load_gcc11.1.sh
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
(again substituting your installation prefix in place of mine). You can then set all these environment variables by simply running:
source load_gcc11.1.sh
Once this is done, you can check that gcc-11.1.0 is properly installed by running:
gcc-11.1 --version
Note that plain old
gcc --version
might still point to an older version of gcc. This is fine though.
###FFTW 3.3.9
Grab the source code for the appropriate versino of FFTW from here: http://www.fftw.org/download.html
However, do NOT follow the installation instructions on the webpage. Those instructions might work if you have root privileges, but I personally couldn't seem to to get things to work that way. Instead, we're going to build fftw with cmake. Untar the fftw source files:
tar -xzvf fftw-3.3.9.tar.gz
Make a build directory and cd into it:
mkdir build
cd build
Now build using cmake, using the flags shown below. For reference, I downloaded and untarred the source file in `/scratch/wluszczak/fftw/build`, so adjust your install prefix accordingly to point to your own build directory that you created in the previous step.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON ../fftw-3.3.9
make install -j $(nproc)
Now comes the weird part. Remove everything except the `include` and `lib64` directories in your build directory (if you installed to a different `CMAKE_INSTALL_PREFIX`, the include and lib64 directories might be located there instead. The important thing is that you want to remove everything, but leave the `include` and `lib64` directories untouched):
rm *
rm -r CMakeFiles
Now rebuild fftw, but with an additional flag:
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/fftw/build/ -DBUILD_SHARED_LIBS=ON -DENABLE_OPENMP=ON -DENABLE_THREADS=ON -DENABLE_FLOAT=ON ../fftw-3.3.9
make install -j $(nproc)
At the end of the day, your fftw install directory should have the following files:
include/fftw3.f
include/fftw3.f03
include/fftw3.h
include/fftw3l.f03
include/fftw3q.f03
lib64/libfftw3f.so
lib64/libfftw3f_threads.so.3
lib64/libfftw3_omp.so.3.6.9
lib64/libfftw3_threads.so
lib64/libfftw3f_omp.so
lib64/libfftw3f.so.3
lib64/libfftw3f_threads.so.3.6.9
lib64/libfftw3.so
lib64/libfftw3_threads.so.3
lib64/libfftw3f_omp.so.3
lib64/libfftw3f.so.3.6.9
lib64/libfftw3_omp.so
lib64/libfftw3.so.3
lib64/libfftw3_threads.so.3.6.9
lib64/libfftw3f_omp.so.3.6.9
lib64/libfftw3f_threads.so
lib64/libfftw3_omp.so.3
lib64/libfftw3.so.3.6.9
Why do we have to do things this way? I don't know, I'm bad at computers. Maybe someone more knowledgeable knows. I found that when I didn't do this step, I'd run into errors that pueoBuilder could not find some subset of the required files (either the ones added by building with `-DENABLE_FLOAT`, or the ones added by building without `-DENABLE_FLOAT`).
Once fftw has been installed, export your install directory (the one with the include and lib64 folders) to the following environment variable:
export FFTWDIR=/scratch/wluszczak/fftw/build
Again, substituting your own fftw install prefix that you used above in place of `/scratch/wluszczak/fftw/build`
###gsl 2.7.1
gsl 2.7.1 is needed for the `mathmore` option in ROOT. If you have an outdated version of gsl, ROOT will still compile, but it will skip installing `mathmore` and `root-config --has-mathmore` will return `no`. To fix this, grab the latest source code for gsl from here: https://www.gnu.org/software/gsl/. Untar the files to a directory of your choosing:
tar -xzvf gsl-latest.tar.gz
For some reason I also installed gsl to my home directory, but in principle you can put it wherever you want.
mkdir /home/wluszczak/gsl
./configure --prefix=/home/wluszczak/gsl
make
make check
make install
To make sure ROOT can find this installation of gsl, you'll again need to set an environment variable prior to building ROOT:
export GSL_ROOT_DIR=/home/wluszczak/gsl/
I also added this to my $PATH variable, though I don't remember if that was required to get things working or not:
export PATH=/home/wluszczak/gsl/bin/:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gsl/lib:$LD_LIBRARY_PATH
###ROOT 6.24.00
Download the specific version of ROOT that you need from here: https://root.cern/install/all_releases/
You might need to additionally install some of the dependencies (https://root.cern/install/dependencies/), but it seems like everything I needed was already installed on my system.
Untar the source you downloaded:
tar -xzvf root_v6.24.00.source.tar.gz
Make some build and install directories:
mkdir build install
cd build
Run CMake, but be sure to enable the fortan, mathmore and minuit2 options. For reference, I had downloaded and untarred the source files to `/scratch/wluszczak/root`. Your installation and source paths will be different.
cmake -DCMAKE_INSTALL_PREFIX=/scratch/wluszczak/root/install/ /scratch/wluszczak/root/root-6.24.00/ -Dfortran=ON -Dminuit2=ON -Dmathmore=ON
Note: if you end up with an error related to compiling XROOTD, then add -Dxrootd=OFF to the original cmake command above.
Then proceed to start the build:
cmake --build . --target install -j $(nproc)
If everything has worked then after the above command finishes running, you should be able to run the following file to finish setting up ROOT:
source ../install/bin/thisroot.sh
##pueoBuilder
By this point, you should have working installations of CMake 3.21.2, gcc-11.1.0, fftw 3.3.9, and ROOT 6.24.00. Additionally, the following environment variables should have been set:
export PATH=/scratch/wluszczak/cmake/install/bin:$PATH
export PATH=/home/wluszczak/gcc-11.1.0/bin:$PATH
export LD_LIBRARY_PATH=/home/wluszczak/gcc-11.1.0/lib64:$LD_LIBRARY_PATH
export CC=/home/wluszczak/gcc-11.1.0/bin/gcc-11.1
export CXX=/home/wluszczak/gcc-11.1.0/bin/g++-11.1
export FC=/home/wluszczak/gcc-11.1.0/gfortran-11.1
export FFTWDIR=/scratch/wluszczak/fftw/build
At this point, the hard work is mostly done. Check out pueoBuilder with:
git clone git@github.com:PUEOCollaboration/pueoBuilder
set the following environment variables:
export PUEO_BUILD_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export PUEO_UTIL_INSTALL_DIR=/scratch/wluszczak/PUEO/pueoBuilder
export NICEMC_SRC=${PUEO_BUILD_DIR}/components/nicemc
export NICEMC_BUILD=${PUEO_BUILD_DIR}/build/components/nicemc
export PUEOSIM_SRC=${PUEO_BUILD_DIR}/components/pueoSim
export LD_LIBRARY_PATH=${PUEO_UTIL_INSTALL_DIR}/lib:$LD_LIBRARY_PATH
Where $PUEO_BUILD_DIR and $PUEO_UTIL_INSTALL_DIR point to where you cloned pueoBuilder to (in my case, `/scratch/wluszczak/PUEO/pueoBuilder`. Now you should be able to just run:
./pueoBuilder.sh
Perform a prayer to the C++ gods while you're waiting for it to compile, and hopefully at the end of the day you'll have a working set of PUEO software.
##Issues I Ran Into
If you already have an existing installation of ROOT, you may still need to recompile to make sure you're using the same c++ standard that the PUEO software is using. I believe the pre-compiled ROOT binaries available through their website are insufficient, though maybe someone else has been able to get those working.
If you're running into errors about c++ standard or compiler version even after you have installed gcc-11.1.0, then for some reason your system isn't recognizing your local installation of gcc-11.1.0. Check the path variables ($PATH and $LD_LIBRARY_PATH) to make sure the gcc-11.1.0 `bin` directory is being searched.
If you're running into an error that looks like:
CMake Error: The following variables are used in this project, but they are set to NOTFOUND.
Please set them or make sure they are set and tested correctly in the CMake files:
FFTWF_LIB (ADVANCED)
then pueoBuilder can't seem to find your fftw installation (or files that are supposed to be included in that installation), try rebuilding with different flags according to which files it seems to think are missing.
If it seems like pueoBuilder can't seem to find your fftw installation at all (i.e. you're getting some error that looks like `missing: FFTW_LIBRARIES` or `missing: FFTW_INCLUDES`), check the environment variables that are supposed to point to your fftw installation (`$FFTWDIR`) and make sure there are the correct files in the `lib` and `include` subdirectories.
|
|
|
157
|
Mon Jun 6 14:19:59 2022 |
Alex M | Important Runs (2) | This is a duplicate post of a previous post from the end of 2020 where I listed the important runs with some of their details in a table (as below). I am extending this table to include the important runs that have been conducted since this post. This includes the run used for the paper as well as the curved run done earlier this year.
In order to access the data for these runs, you can find them by going to this directory: /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs
Some of these runs can also be accessed in the old project space directory, though they should all be contained in the above directory. Here's the path if interested:
The runs are contained in directories available in the above path. Use caution when listing files in some of these directories--some contain many files (primarily the .uan files -- more recent runs are better organized), which means it may take a long time to list files/directories.
| Name |
Description |
Symmetry |
NPOP |
Generations |
Roulette/Tourney/Rank |
Crossover |
Reproduction |
Mutation |
Injection |
Penalty |
Neutrinos |
|
| Machtay_20200824_Real_Run |
First real run with significant amounts of data after the summer improvements.______________________________________
|
Symmetric |
10 |
15 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
100k |
|
| Machtay_20200827_Asym_Length_Run |
First asymmetric length run after summer improvements. |
Asymmetric length |
10 |
17 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
100k |
|
| Machtay_20200831_Asym_Length_and_Angle |
Asymmetric length and angle run after summer improvements. |
Asymmetric length and angle |
10 |
42 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
150k |
|
| Machtay_20200911_Symmetric |
Longer symmetric run with fewer neutrinos. |
Symmetric |
10 |
35 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
30k |
|
| Machtay_20200914_Asymmetric_50_Individuals |
Longer asymmetric run with fewer neutrinos. |
Asymmetric (all dimensions) |
50 |
26 |
100% Roulette |
100% |
0% |
- |
100% |
Yes |
30k |
|
| Machtay_20201016_Symmetric_Improved_GA |
First run using improvements to GA based on Ryan's paperclip/fast loop analysis. |
Symmetric |
50 |
10 |
50/50/0 |
75% |
10% |
- |
15% |
Yes |
30k |
|
| Machtay_20201023_300K_Nus_50_Individuals |
Started with all identical individuals to demonstrate evolution; replaced penalty with hard cutoff. Increased Nus for higher fitness score precision. |
Asymmetric (all dimensions |
50 |
25 |
50/50/0 |
75% |
10% |
- |
15% |
No |
300k |
|
| AraSim_Polarity_Fix_2021_03_19 |
Run used in the paper. In this run, we fixed an error that had been noticed by Brian and Jorge in AraSim. The error involved the polarity of the signals in Report.cc (hence the name of this run). |
Asymmetric(all dimensions) |
50 |
31 |
80/20/0 |
72% |
6% |
- |
22% |
No |
300k |
|
| 2022_02_08_Rank_Test |
This was the first long run done using a new gene for the curvature of the cones. We recast the side lengths to be described by the coefficients of a quadratic polynomial, rather than by the opening angle. This also used rank selection instead of roulette.
Additionally, mutation has been changed here so as to apply small perturbations to existing genes rather than regenerating those genes altogether. This only applies to individuals created by crossover. The mutation column indicates the probability of mutating a gene and the standard deviation of the gaussian that determines the change (in terms of % of the original value).
|
Asymmetric, Quadratic |
50 |
50 |
0/90/10 |
72% |
6% |
1%, 5% |
22% |
No |
300k |
|
| 2022_04_14_Identical_Asym_Lower_Min |
This run used the asymmetric GA to see if by lowering the minimum length (down to 10 cm instead of 37.5) the GA would try to run away to ever smaller lengths. |
Asymmetric |
50 |
6 |
2/8/0 |
72% |
6% |
- |
22% |
No |
300k |
|
|
|
|
124
|
Mon Dec 7 22:51:08 2020 |
Alex M | Important Runs | Today I removed some of the run directories which had very little or no data or weren't worth keeping around. There are still a few that I think can be removed, but I'm keeping them until we can get a consensus that they can definitely be removed. Below I listed the names and descriptions of the runs that I think we should definitely preserve going forward. In general, the more data contained in the run directory, the more important it is to keep around.
| Name |
Description |
Symmetry |
NPOP |
Generations |
Roulette/Tourney |
Crossover |
Reproduction |
Mutation |
Penalty |
Neutrinos |
|
| Machtay_20200824_Real_Run |
First real run with significant amounts of data after the summer improvements.______________________________________
|
Symmetric |
10 |
15 |
100% Roulette |
100% |
0% |
100% |
Yes |
100k |
|
| Machtay_20200827_Asym_Length_Run |
First asymmetric length run after summer improvements. |
Asymmetric length |
10 |
17 |
100% Roulette |
100% |
0% |
100% |
Yes |
100k |
|
| Machtay_20200831_Asym_Length_and_Angle |
Asymmetric length and angle run after summer improvements. |
Asymmetric length and angle |
10 |
42 |
100% Roulette |
100% |
0% |
100% |
Yes |
150k |
|
| Machtay_20200911_Symmetric |
Longer symmetric run with fewer neutrinos. |
Symmetric |
10 |
35 |
100% Roulette |
100% |
0% |
100% |
Yes |
30k |
|
| Machtay_20200914_Asymmetric_50_Individuals |
Longer asymmetric run with fewer neutrinos. |
Asymmetric (all dimensions) |
50 |
26 |
100% Roulette |
100% |
0% |
100% |
Yes |
30k |
|
| Machtay_20201016_Symmetric_Improved_GA |
First run using improvements to GA based on Ryan's paperclip/fast loop analysis. |
Symmetric |
50 |
10 |
50/50 |
75% |
10% |
15% |
Yes |
30k |
|
| Machtay_20201023_300K_Nus_50_Individuals |
Started with all identical individuals to demonstrate evolution; replaced penalty with hard cutoff. Increased Nus for higher fitness score precision. |
Asymmetric (all dimensions |
50 |
25 |
50/50 |
75% |
10% |
15% |
No |
300k |
|
| |
|
|
|
|
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
152
|
Fri Apr 8 16:07:22 2022 |
Alex M | Identical Asymmetric Lowered Length Minimum Run | We are trying to do a short run of the asymmetric bicone so that we can see how it tries to evolve the antenna when the minimum length is lowered from 37.5 cm to 10 cm. Currently, there is a problem with the loop in the asymmetric version. Attached is the run detail file. |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2022_04_07_Identical_Asym' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=30000 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=0 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=2 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=8 ## Percent of individuals selected through tournament (divided by 10)
RANK=0 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
#####################################################################################################################################################
|
|
|
184
|
Mon Oct 24 17:44:51 2022 |
Ryan Debolt | Icemc inputs | Here is our current assumption of the antenna angles needed for the icemc inputs. |
| Attachment 1: 261D6FA3-5612-412F-80FA-8AD961F7331A.jpeg
| 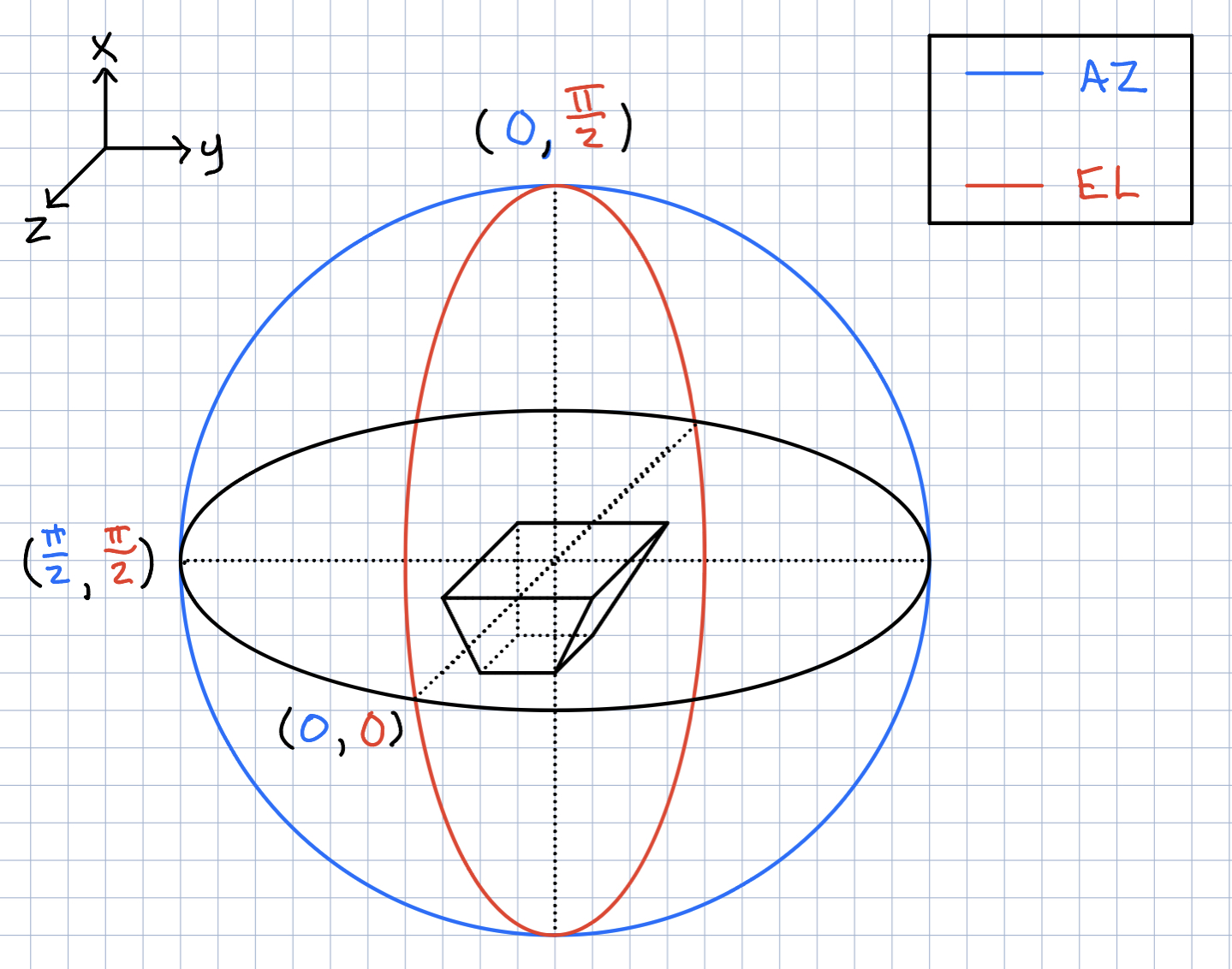 |
|
|
182
|
Fri Sep 2 12:40:17 2022 |
Alan S | Ice Shell Around Bicone | Here is a PDF with a summary/log of my work trying to integrate an ice shell surrounding the bicone antennas in the GENETIS loop. I made gain patterns of the ARA bicone with the ice shells and without them, showing the impact of the ice on the antenna's gain. Having an ice shell changes these patterns, which are also dependent on the radius of the shell. I showed that convergence on the gain patterns and expected features start ocurring for shells with radii larger than 10 meters. I also keep track of the computation time which increases with bigger shells.
Note: I also add PowerPoint slides that contain all the graphs that I made. The PDF does not contain some of those since I didn't think they were coherent with my write-up, but I may be making an omission.
|
| Attachment 1: XF_ARA_bicone_in_ice_and_air_column.pptx
|
| Attachment 2: Far_Zone_Gain_of_ARA_Bicone_insice_Ice.pdf
|
|
|
217
|
Wed May 10 17:43:20 2023 |
Alex M | How to run the loop | In this post, I'm going to give step by step instructions on everything you need to do to run the loop. See Julie's thesis for the most recent iteration of this manual. Also refer to entry 123 for a list of some errors we have encountered historically in the loop and how to resolve them, though some fixes may be outdated.
Running the loop
Getting started
Assuming you have followed the instructions from the onboarding tutorials, you should be ready to access OSC through the interactive website. Instead of using ssh to access OSC, the loop requires that you go to this website: https://ondemand.osc.edu/ . Login with your usual OSC credentials. Once there, you will need to request a Lightweight Desktop by clicking on "Interactive Apps" at the top of the page. See the screenshots attached for how to do this. Old documentation may refer to the Lightweight Desktop as a VDI.
You can request up to 24 hours for a Lightweight Desktop, and it should begin almost immediately. After you request it, the website will take you directly to the waiting page. You can navigate to this page yourself by clicking "My Interactive Sessions" at the top of the ondemand page. When the job begins, you will see a blue buttong that says "Launch Lightweight Desktop". Click it and you should have a new tab open that brings you to a remote desktop environment on OSC that you interact with through your browser.
New runs
If you're starting a new run for the first time (that is, beginning from generation 0), you will need to edit the script that runs the loop. Here is the path: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Loop_Scripts/Asym_XF_Loop.sh . At the top of the script is a list of variables that can be changed. See the first screenshot attached here.
For a new run, you must change the variable <RunName>, which is the first variable in the list. It should be in quotes and the convention is to start the name off with the date of the run, using YYYY/MM/DD format--this will always keep everything in chronological order when listing directories with <ls>. Other variables are explained in the comments next to them in Asym_XF_Loop.sh and you should change them depending on the type of run you are conducting.
To begin the loop, type in the command ./Loop_Scripts/Asym_XF_Loop.sh from the Evolutionary_Loop directory.
When the loop begins, you will be asked to press any key. After you hit a key, the loop will run Part A, which executes the GA using the <start> mode. It will then automatically move on to Part B, which creates the antennas in XFdtd. XF will ask you if you want to specify a location for the XF file to be saved. Click <No>. The loop will automatically save the XF directory into the run directory when it finishes with the first part of part B. In principle (that is, barring errors that stall the loop or interruptions due to the VDI job ending), you will never need to manually enter anything to the loop for it to run beyond this point.
However, if it is your first time using XF, you will need to set the max GHz setting upon startup to 2 instead of the default 10.
Continuing Runs
After starting a new run, the loop will run automatically. When your Lightweight Desktop job ends, you'll need to get another one repeat the steps above. You will not be prompted for any input.
Stepping Back
At times, you may want to continue the loop from an earlier point or interrupt the loop and step back. Usually you'll only want to do this within a generation, but it's possible that you will encounter an error or bug at the end of a generation and wish to return back to that point to remedy it before continuing forward in the next generation. This is a very tricky and delicate process that requires a lot of attention to detail. In the future, we'd like to make an option for the loop to do this automatically when given input at the beginning, but implementing that will take a long time and will be very sophisticated.
The state of the loop is recorded in the savestate file located here: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/saveStates . The file will be named <Run_Name>.savestate.txt . In it, you will see three numbers. The first one represents the current generation of the loop. The second one represents the state of the loop, that is, which part in the generation it is currently on. The third one represents the individual it is processing. In practice, the third number is rarely relevant, and is almost always 1, though it can be used for Part B, where the loop is processing individuals sequentially, so if it was interrupted on individual eight (for example) you can change the number to be 8 so that it will pick up where it left off without needing to redo the first seven individuals.
If you want to step back to an earlier state in a generation, you'll need to interrupt the loop and revert the state by changing the second number in the savestate file. This is NOT all you will have to do, however. Because of the way files are edited and moved around during states, you will need to manually revert files to be in the format or location needed by the earlier state.
For example, you may need to revert from state 2 (when XF is creating the antenna geometries) to state 1 (when the GA generates new individuals). In this case, you'll need to edit the savestate file to change the second number from a 2 to a 1. Then, you'll need to change the generationDNA.csv file in Generation_Data to use the values from the previous generation, since the GA will have overwritten that file and uses it to determine what individuals are to be selected from. To do this, you'll need to locate the <gen>_generationDNA.csv file from the previous generation in the <RunName> directory in Run_Outputs. This is should be found here: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/<RunName>/Generation_Data .
The above is a relatively simple example of how to step back in the loop. Stepping back from a late state to an early state will be more complicated, though if you're willing to sacrifice run time you can pretty easily follow the above to just restart a generation. Stepping back to a previous generation will be explained in a future update to this post. |
| Attachment 1: Ondemand_Front_Page.png
|  |
| Attachment 2: Ondemand_Interactive_Apps.png
|  |
| Attachment 3: Launching_Lightweight_Desktop.png
|  |
|
|
177
|
Tue Aug 9 15:28:50 2022 |
Ryan Debolt | How many individuals to use in the GA. | One of our foundational questions tied to the optimization of the GA has been "How many individuals should we simulate". Up to now, our minds were made up for us by the speed of arasim being great enough that the time cost of simulating individuals was great enough that the improvements made from having more were not enough to justify the slowdown. However, with the upgrade to the faster, more recent version of arasim, I decided to re-examine this. This was also spurred on by the fact that the last time I ran this test we were testing GA performance by final generation metrics rather than by how many generations it took to reach a benchmark. So in one of my optimization tests, I tracked this data.
To start, using the same run proportions, using a .5 chi-squared benchmark, the average time across all 89 run types used in this run was 25.4 generations for 50 individuals as compared to 8.3 generations running the same test for 100. Furthermore, the minimum number of generations for 50 individuals was 4.8 while using 100 individuals yielded 2.4. So on average running 100 individuals was about 3 times fast at reaching this benchmark than with 50. And when comparing the best result regardless of run type, 100 individuals was still 2 times quicker than the min for 50 individuals. Finally, the run that yielded an average of 2.4 generations for 100 individuals took an average of 29.2 generations with 50 individuals or roughly 12 times the generations.
For the test we will discuss, we ran 89 different run types that all used 60% rank, 20% roulette, and 20% tournament selection respectively. These test had the following ranges:
6-18% of individuals through reproduction (steps of 3%)
64-88% of individuals through crossover (steps of 12%)
0-10% mutation rate (steps of 5%)
1-5% sigma on mutation (steps of 1%)
These tests also used our fitness scores with simulated error of .1 to imitate arasim's behavior and as such we used the chi-squared value to evaluate these scores as there is no error on those values.
Comparing this same test with a tighter chi-squared benchmark of .25, we see similar results. On average 50 individuals took 37.1 generations to reach this point while 100 individuals took 16.0 generations. Similarly, the minimums amount of gens for 50 individuals was 15.4 while 100 individuals was 5. Finally, the corresponding run for the 5 generation min with 100 individuals took 41.8 generations with 50 individuals. These correspond to speed up's of 2.3, 3.08, and 8.36 respectively.
This data implies that on average, independent of run type, we should expect to have to use 2-3 times fewer generations while running 100 individuals than we would running 50 individuals but we could see up to 8-12 times fewer generations to reach benchmarks. Another data set using a different set of selection methods was also tested for this and again yielded similar results, though overall the runs from the first batch were better across both 50 and 100 individuals and so those results are likely to be more indicative of the parameters we use in a true run.
The data being examined in these results can be found here: https://docs.google.com/spreadsheets/d/1GlfnjQSO6VI8MuUGYTUcLkjwDZU98nyFFysgTTfVFOE/edit?usp=sharing |
|
|
210
|
Wed Apr 5 17:21:07 2023 |
Nick King | H-pol XF Design | Work in progress. Update on H-pol XF design. The first plot is the gain pattern with ferrite rod sets (blue) vs. with nothing but the copper plates.
The second plot is the gain pattern with everything but the ferrite rod sets vs. with nothing but the copper plates.
The ferrite rod sets function to narrow the gain pattern. |
| Attachment 1: Screenshot_(7).png
| .png) |
| Attachment 2: Screenshot_(8).png
| .png) |
| Attachment 3: HpolfromScript(5_10_2023).png
| .png) |
| Attachment 4: StephHpolwith_just_copper_plates_and_wires.png
|  |
|
|
222
|
Thu Jun 1 21:49:29 2023 |
Dylan Wells | Guide to Updating pueoSim | How To Update PueoSim For GENETIS:
First, whoever updates pueoSim needs access to pueoBuilder, pueoSim, and niceMC on GitHub (ask Will for permissions).
Once you do, go into the peuoSim directory at /fs/ess/PAS1960/buildingPueoSim/
and source set_env.sh
`source set_env.sh`
Then, we want to make copies of the files we are currently modifying for the GENETIS loop.
For PueoSim v1.1.0 these are:
-
nicemc/src/EventGenerator.cc # Modified to create custom output root files to calculate errors
-
pueosim/test/simulatePueo.cc # Modified to stop the simulation of the LF antenna which is not needed for GENETIS
-
pueosim/src/Seavey.cc # Modified to read in custom gain files
-
pueosim/src/pueo.cc # Modified to read in custom gain files
-
pueosim/include/Seavey.hh # Modifies to read in custom gain files
Currently, I store copies of these in the `/fs/ess/PAS1960/buildingPueoSim/backups` directory in case somebody accidentally overwrites the files in pueoBuilder.
Once you’ve made the copies, you can run `./pueoBuilder.sh` from the `/fs/ess/PAS1960/buildingPueoSim/pueoBuiler` directory. This will rebuild pueoSim and niceMC, pulling the latest updates from GitHub.
You may need to delete the pueoSim and niceMC directories in order for the builder to pull the latest version from GitHub. Or, if it’s being really stubborn, move the whole pueoBuilder directory to a temporary location and run the builder from scratch with
`git clone git@github.com:PUEOCollaboration/pueoBuilder` and then `./pueoBuilder.sh`
Then, you will need to reference the copies of the changed files to make changes to the new version of pueoSim. Hope this doesn’t cause too much of a headache, and when you’re done, return to the /fs/ess/PAS1960/buildingPueoSim/pueoBuiler directory.
Then you simply type
`make install`
then
`make`
And now, pueoSim should be ready to run!
EventGeneratior.cc Changes and Rationale:
PueoSim’s default output ROOT files are very large and therefore time-consuming to parse through to get the information we need to calculate effective volume and errors. So, we want to create a custom ROOT file with only the variables we want, greatly increasing the speed of the analysis.
To do this, we want to create a new TFile with corresponding TTree and TBranches that will store the loop.positionWeight, loop.directionWeight, and neutrino.path.weight. Then, we want to fill the Tree when a detector is triggered and write the results to the file at the end of the loop.
Sample code for initializing the TFile:

simulatePueo.cc Changes and Rationale:
By default, pueoSim v1.1.0 runs a simulation for the normal antenna and a low-frequency (LF) antenna. As GENETIS is evolving for the main antenna, we are not interested in using computation time to simulate the LF antenna. So, we comment out the lines that initialize the LF detector in this file.
Seavey.cc, Seavey.hh, and pueo.cc Changes and Rationale:
As of pueoSim v1.1.0, the simulation software only has the ability to read in one antenna. This is a problem, as we want it to be able to read in files for any antenna we make. So, we need to change these scripts to be able to read in whatever gain files we want.
The convention the loop uses is to input gain files in the format of
vv_0_Toyon{runNum}
hh_0_Toyon{runNum}
…
In the ./pueoBuilder/components/pueosim/data/antennas/simulated directory
So, the main change is getting the runNum variable into Seavey.cc file where the gains are read. Currently, we define a global variable at the stary of pueo.cc and pass it into where Seavey is called. This means we have to add runNum as a parameter in Seavey.cc as well as Seavey.hh. Finally, we change the name of the read-in files to be in the above format AFTER dividing runNum by 1000 (this is because pueoSim uses the run number as a random number generating seed, so we divide runNum by 1000 to be able to read in the same gain patterns for multiple seeds of the same individual).
Note: We used to change pueoSim to output a veff.csv file. We now handle this calculation by analyzing ROOT files, so this change is no longer necessary. |
|
|
123
|
Wed Dec 2 15:24:39 2020 |
Alex M | Guide for Loop Errors | Attached is a .txt file you can find in the Evolutionary_Loop directory as Loop_Error_List.txt. It's a list of the current errors we sometimes experience in the loop, along with how to fix them if you encounter them while running. If people encounter errors that aren't in the list, let everyone know in the #genstudents chat on slack and update the file with the error message and when it was encountered (what state the loop was in) and the possible cause and solution if you know it. |
| Attachment 1: Loop_Error_list.txt
|
Below is a list of errors we may encounter in the loop as of 11/25/20:
Error: "Pre-while, pre-for"
Description: This is an error you'll encounter after AraSim has "completed." The loop will hang after outputting "Pre-while" and then "Pre-for." This comes from the fitness function--the loop is indicating that it is inside the fitness function right before it enters the two loops it runs over the AraSim data. Hanging here indicates that there was an issue running AraSim. Specifically, it indicates that at least one of the jobs for the *first* individual in AraSim failed.
Potential causes:
This may be caused by an issue in generating the gain files used for AraSim. These gain files are placed in the AraSim directory under the names a_{num}.txt, where {num} represents the individual number. You can check a_1.txt in the AraSim directory and see if it's complete (if it isn't, you can usually tell just by opening the file and seeing that only two lines have been printed to it).
One way of looking for the cause of this error is to look at the job error files. Inside the {runname} directory are directories containing the error and output files from the AraSim jobs. These are .../{gen}_AraSim_Errors and .../{gen}_AraSim_Output, where {gen} is the generation number. One example of an error message I've seen in the error files was
"
terminate called after throwing an instance of 'std::out_of_range'
what(): basic_string::substr
/var/spool/slurmd/job2345909/slurm_script: line 37: 171196 Aborted (core dumped) ./AraSim setup.txt $runNum outputs/ a_${num}.txt > $TMPDIR/AraOut_${gen}_${num}_${Seeds}.txt
"
This appeared in all of the error files for the first individual's jobs.
Resolution:
The best way to resolve this is to start by checking the error files. In the case of the error message above, it would be best to go to the AraSim directory and check for the a_{num}.txt file. If you see just one (ex: a_1.txt), then that's likely to be the culprit (especially if the file is obviously not completely filled)--these files should be removed, so if one is left it may not have been moved correctly, likely due to permissions errors. Remove the a_{num}.txt file and restart from the AraSim job submissions (potential speed up: add part of the error message to the self-correcting phrases in Part_D2_AraSeed.sh to only rerun those individuals).
It's also possible that the issue was caused in XF. Make sure you follow the instructions above and start back at stage 2 to restart from the beginning of XF if starting back from AraSim doesn't work. Try to take notes on the differences you see to add to this.
It's also notable that this may be caused by permissions issues. Every time someone is handing the loop off to someone else, the OpenPermissions.sh script should be run (passing the {runName} as an argument). Look in that script to determine which files need to have open permissions. If the person with ownership of the closed files isn't available to open them, you can remove them and start back from where they would have been created. This usually occurs in AraSim and the AraOut files in Antenna_Performance_Metric should have their permissions fixed or be removed (for that generation only).
*****************************************************************************
Error: <Loop hangs while outputting dimensions and fitness scores>
Description:
This is similar to the error above, except that instead of hanging on the first individual, it hangs on some later individual.
Resolution:
The instructions for resolving this should be the same as the ones above. This seems to be less common and is usually resolved by the self-correcting code in Part_D2. Regardless, if you encounter this error the first step should be to follow the instructions below in case there is just one or a handful of failed AraSim jobs. If that doesn't work, step back to stage 5 to resubmit the AraSim jobs after clearing out the possible offending files. If that doesn't work, step back to XF (you can always just step back to XF at the beginning if you're unsure that stepping back to AraSim will resolve this to potentially save time).
It's also possible that there is an error in just a handful (or even just one) of the AraSim jobs. This might be caused by opening permissions after someone has already taken over running the loop. In this case, you might be able to start the loop back up without needing to resubmit all of the AraSim jobs or step back all the way to XF. To do this, you'll need to figure out which AraSim job failed. Check the AraSim error files and output files for that generation (specifically, check to see if one is *missing*). You should be able to figure out which individual the loop is stuck on by counting how many sets of dimensions and fitness scores were printed to screen before the loop started hanging. Go to /Antenna_Performance_Metric (inside .../Evolutionary_Loop) and list all of the AraOut files corresponding to that individual and check them to see if any of them appear incomplete (AKA don't have an effective volume at the bottom).
Once you find the individual jobs that failed, you can set up the loop to only rerun those jobs. First, go to the AraSim flags directory inside the RunName directory and populate the the flags like so:
>for i in `seq 1 <NPOP>
>do
>for j in `seq 1 <jobs per individual>
>do
>echo <gen> > ${i}_${j}.txt
>echo $i >> ${i}_${j}.txt
>echo $j >> ${i}_${j}.txt
>done
>done
This will populate all of the flag files needed for AraSim to move on. Remove the flag files corresponding to the identified failed AraSim jobs. Next, go into the AraSim error file directory in the RunName directory (/<gen>_AraSim_Errors) and replace any text inside the error files corresponding to the failed AraSim jobs with the phrase "segmentation violation" (spelling and capitalization matter!). This is one of the phrases used in the self correcting part of the loop in Part_D2 and indicates to the loop to resubmit the AraSim job for that individual.
After doing this, you should be able to return to .../Evolutionary_Loop and change the savestate in /savestates to 6 from 7. Now you can start back the loop and it will tell you that it's waiting for the AraSim jobs to finish. After 1/2 minutes it will notice that the error files have "segmentation violation" in them and will resubmit only the AraSim jobs you specified as having failed.
*****************************************************************************
Error: "cannot connect to X11 forwarding" (or something to that effect)
Description:
This usually occurs during XF, but it may occur during the display of plots. In the case of plots being unable to display, the loop should still be able to operate, though plots might not update. However, if this message occurs during XF the data for the gain patterns for the antennas won't be properly created. This can occur at the first opening of the XF GUI or on the second part (after the xfsolver jobs have run).
Potential causes:
First, you should make sure that you are logged in to OSC using <ssh -XY userID>. The -XY allows x11 forwarding, which is needed for the XF GUI to appear. Also remember to indicate that you need X11 forwarding when requesting your interactive job (using --x11). It's also possible that your connection to the X11 forwarding can be interrupted after a long time (I've seen the loop work for several generations over multiple interactive job submissions and then suddenly get this error).
Resolution:
My advice is to log out and back in to OSC each time your interactive job ends. This is an uncommon error but it's easy to miss. Once you've logged back in, you'll need to restart the save state back to the part of XF where you got this error (either 2 or 3 depending on which par the error appeared in).
*****************************************************************************
Error:
|
|
|
224
|
Mon Jun 5 15:00:07 2023 |
Ryan Debolt | Github tests | We want to automate tests in pull requests in GitHub to run unit tests on individual functions to ensure they return the expected results. This will help prevent bugged code from being pushed to working branches. We will add a list of unit tests to this ELOG as we design them.
|
|
|
228
|
Thu Jun 29 12:05:45 2023 |
Alex M | Gain at various source heights | Here are a collection of antenna gain patterns with the antenna power source located at various heights at 500 MHz. The heights are indicated in the name of the images and were located at 3cm, 6cm, 12cm, and 24cm from the base. One thing to note is that the power source becomes longer as a function of height (since it's connected to the ridges, which slope back).
The 3D gain plots attached show the gain becoming more uniform until we get to the last one, where it seems to reinvert and be much stronger in the upward direction than below. I'll update this post with some polar slices for a more complete comparison between them.
EDIT: 07/17/23
I've made a few adjustments and added plots of them.
- First, I separated the ridges. Previously, the ridges touched at their inner corners. This electrically connected them and may be why we saw very high peaks in the first set of impedance plots. You can see the plots named Separated_Impedance_<x>_cm.png.
- This appears to have lowered the peaks considerably and also shifted them to lower frequencies. The peaks are still high when the source is placed high.
- We seem to be developing another peak at higher frequencies, although this also existed in the previous tests for the high sources.
- After this, I added a waveguide and positioned the source between the extensions of the ridges into the waveguide. I've also added a picture that shows the waveguide in the geometry.
- The sources here are positioned from 1 cm below the ridges to 8 cm below the ridges. The naming convention is the same as above, except it includes "Waveguide" in the name.
- The peaks are considerably lower here and seem consistent as we place sources further down. They are all below 500 Ohms, compared to as much as 4000 Ohms in the previous tests.
EDIT: 08/10/23
I've been trying more tweaks to the design to get rid of the peaks we see in the plots below. I'm having trouble actually getting the impedance to match close to the 50 Ohm value we expect from the cable. The best case situations seem to come from when the ridges are still touching, which we don't want. Here are the properties I've been changing:
- Ridge positions (gene in GA)
- We don't want the ridges to touch, but I can't get them to be very far from each other anyway. The minimum wavelength in our bandwidth would be 20 cm, but that's close to the size of the bottom of the antenna for when we make it large, so the separation is still small.
- When the ridges do touch is actually when I get the best matching,
- Bottom size (gene in GA)
- I've been changing the size of the bottom of the antenna. This is something that can change substantially between individuals in the GA, so even if I find a good value for this, the impedance it gives wouldn't be the same for all individuals we see in an evolution.
- This does seem to move the location of the impedance peak (in frequency) but I can't seem to get it fully out of band.
- Waveguide length
- I've been changing the length of the waveguide between 3 cm and 10 cm. We said that somewhere in the middle should be the most reasonable (the example antenna we saw looked to have a waveguide of ~5 cm long). It seems like the source position is more important.
- Source position
- I've been moving the position of the power source inside the waveguide. This seems to move around the peak and can lower its height, but I haven't been able to get peaks lower than ~400 Ohms, except in cases where the ridges are in contact.
I've added a picture of the best impedance I managed to get, but this involved an antenna with a small base and ridges touching. My next step is to just try a grid search by making many antennas automatically and simulating them all to look for any that have a nice impedance in our band. |
| Attachment 1: 3_cm.png
| 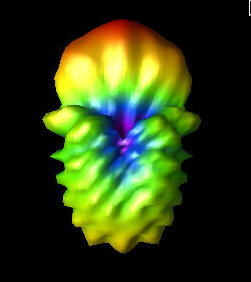 |
| Attachment 2: 6_cm.png
| 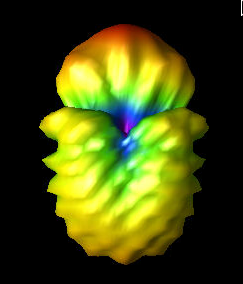 |
| Attachment 3: 12_cm.png
| 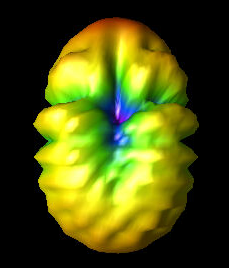 |
| Attachment 4: 24_cm.png
| 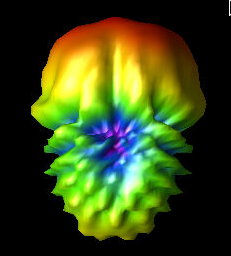 |
| Attachment 5: PUEO_Impedance_3_cm.png
| 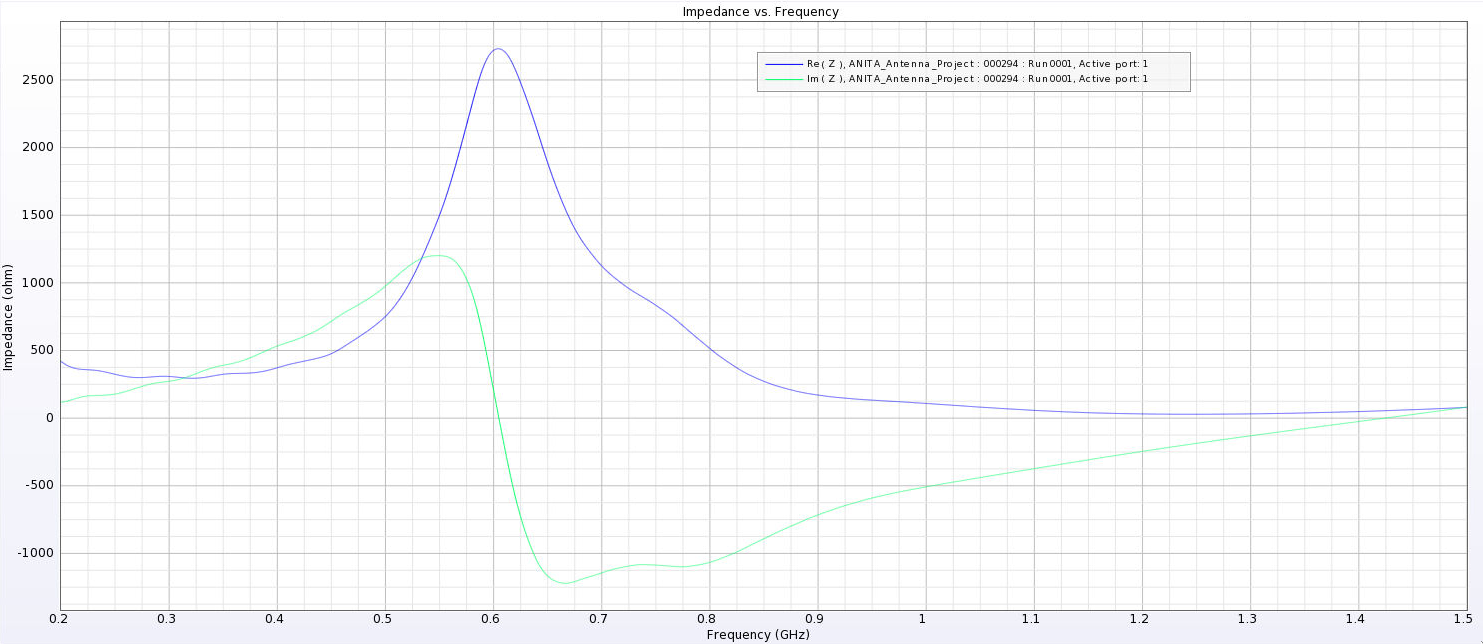 |
| Attachment 6: PUEO_Impedance_6_cm.png
| 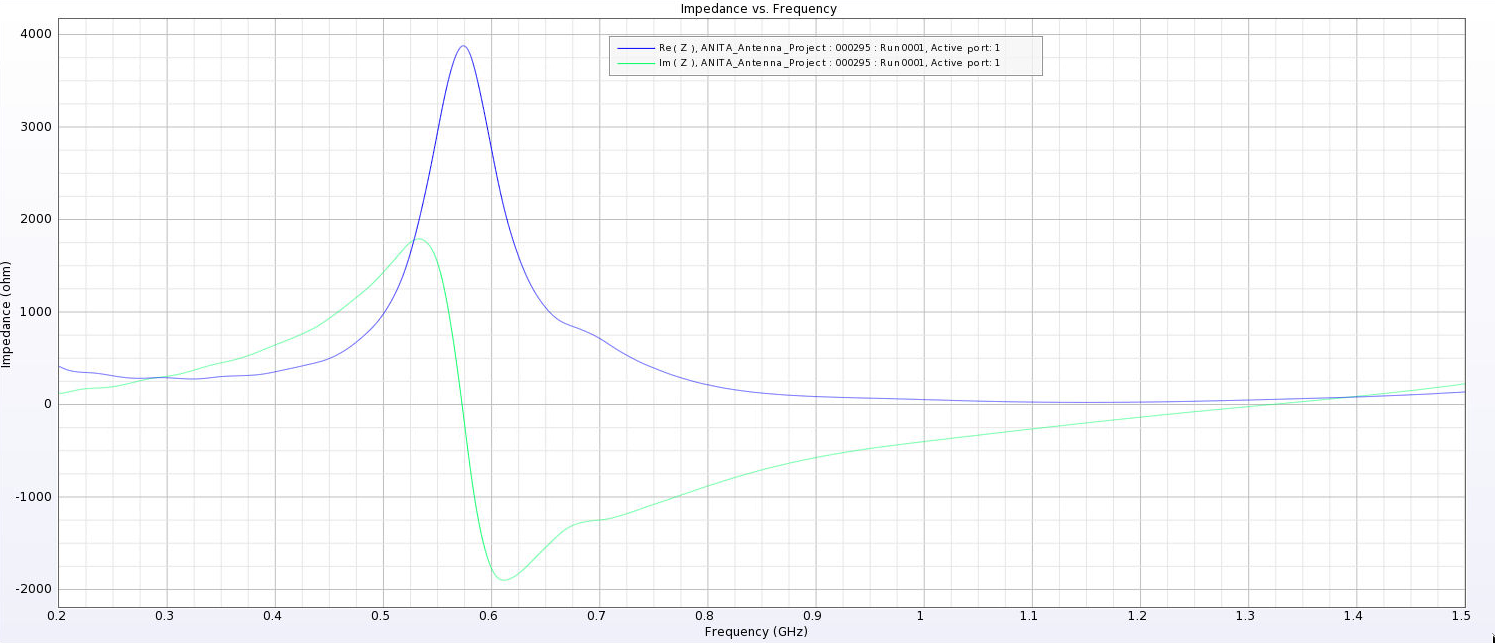 |
| Attachment 7: PUEO_Impedance_12_cm.png
| 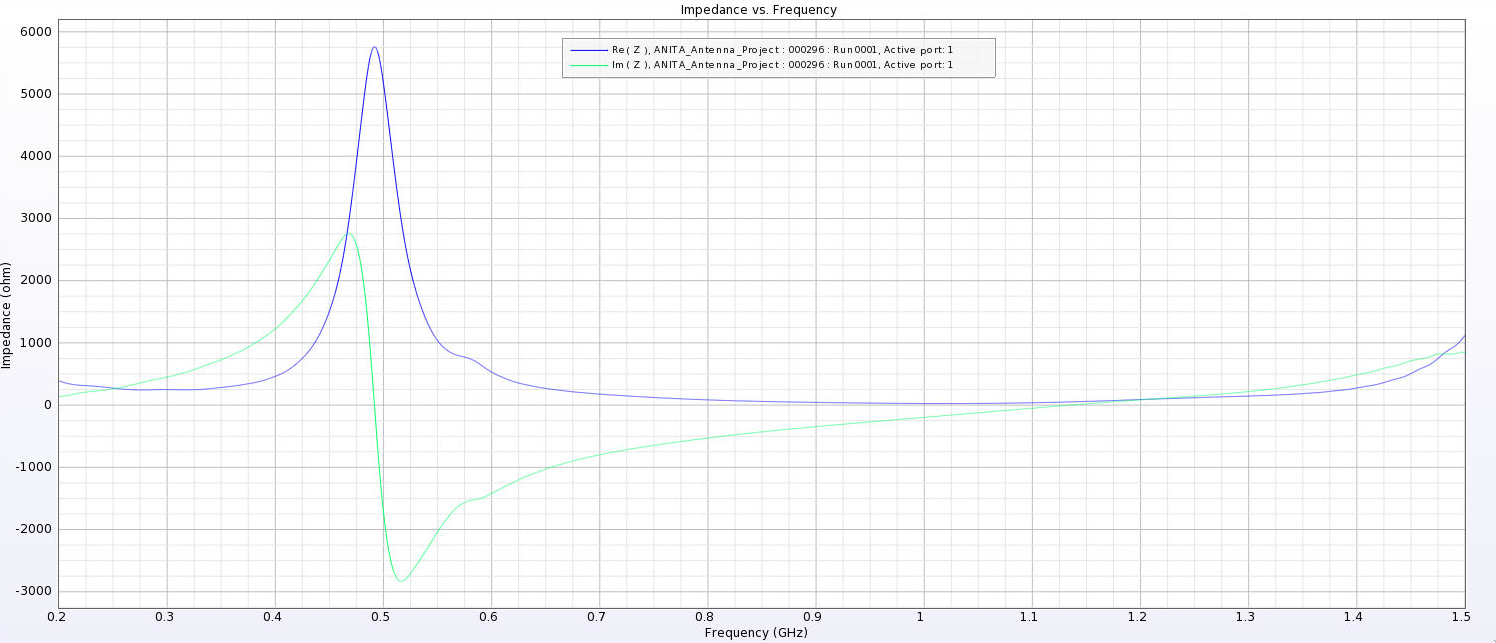 |
| Attachment 8: PUEO_Impedance_24_cm.png
| 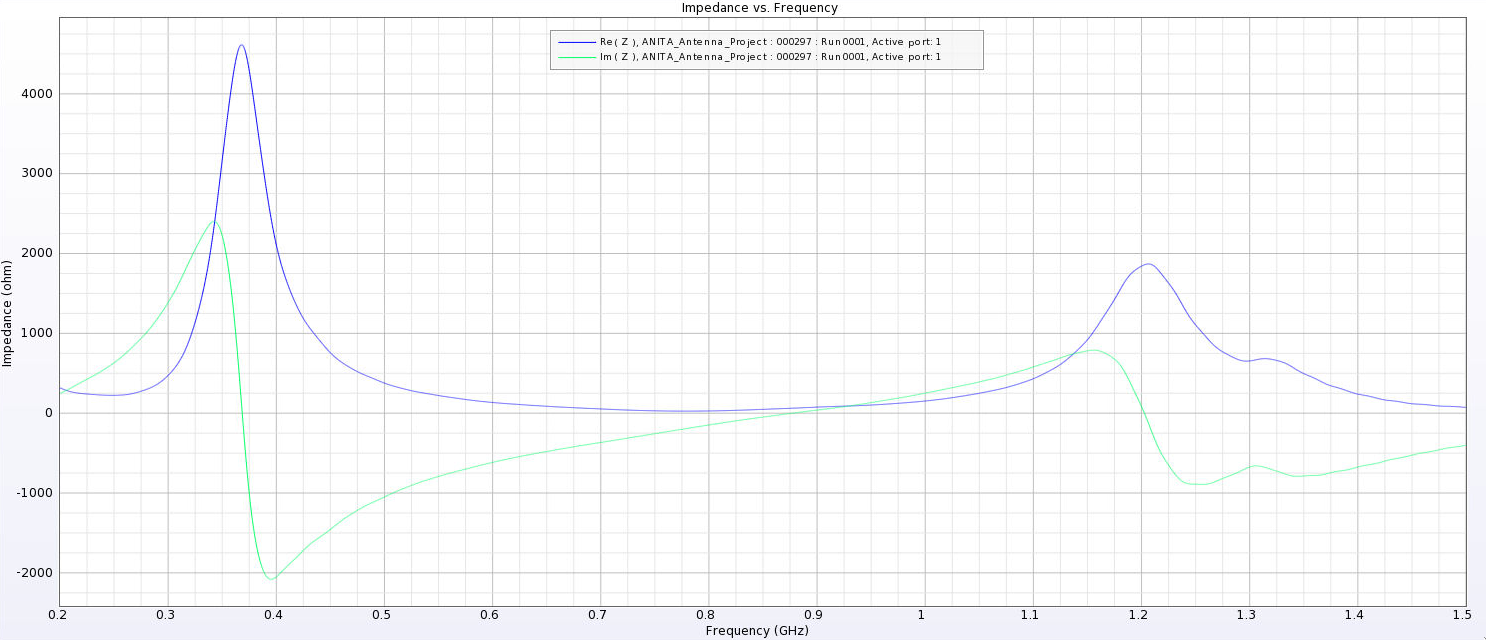 |
| Attachment 9: Separated_Impedance_3_cm.png
| 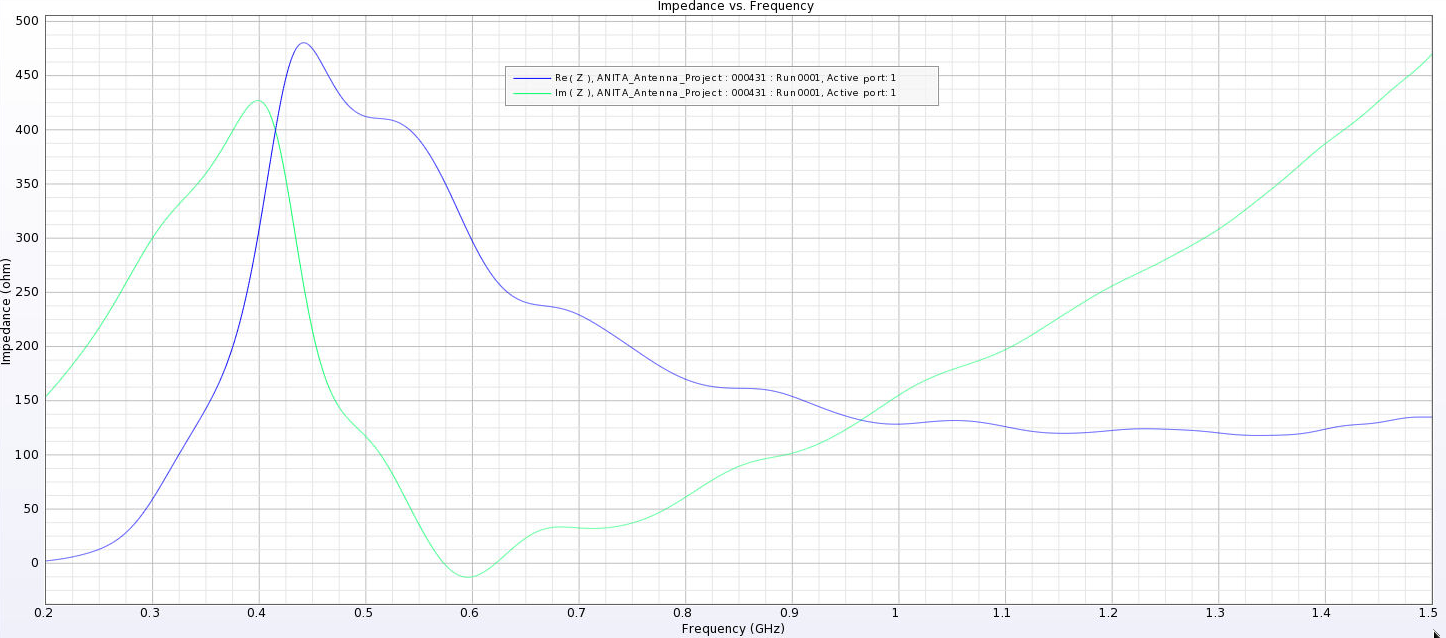 |
| Attachment 10: Separated_Impedance_6_cm.png
| 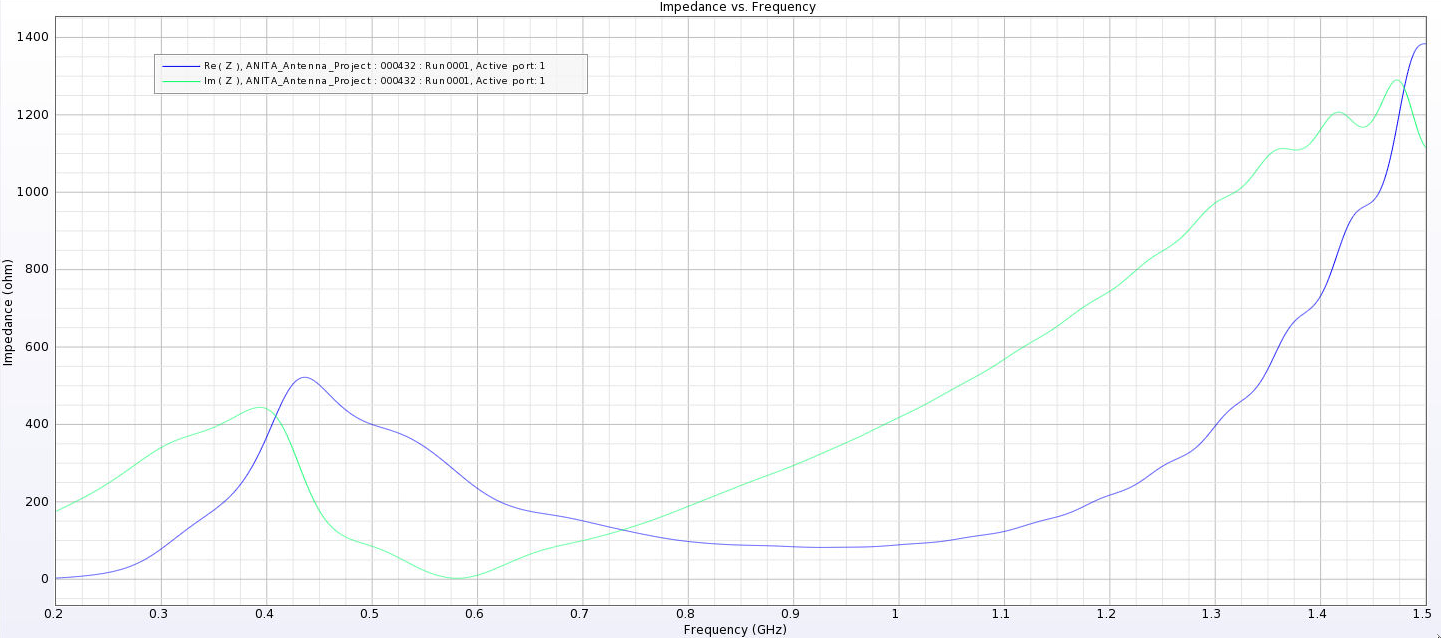 |
| Attachment 11: Separated_Impedance_12_cm.png
| 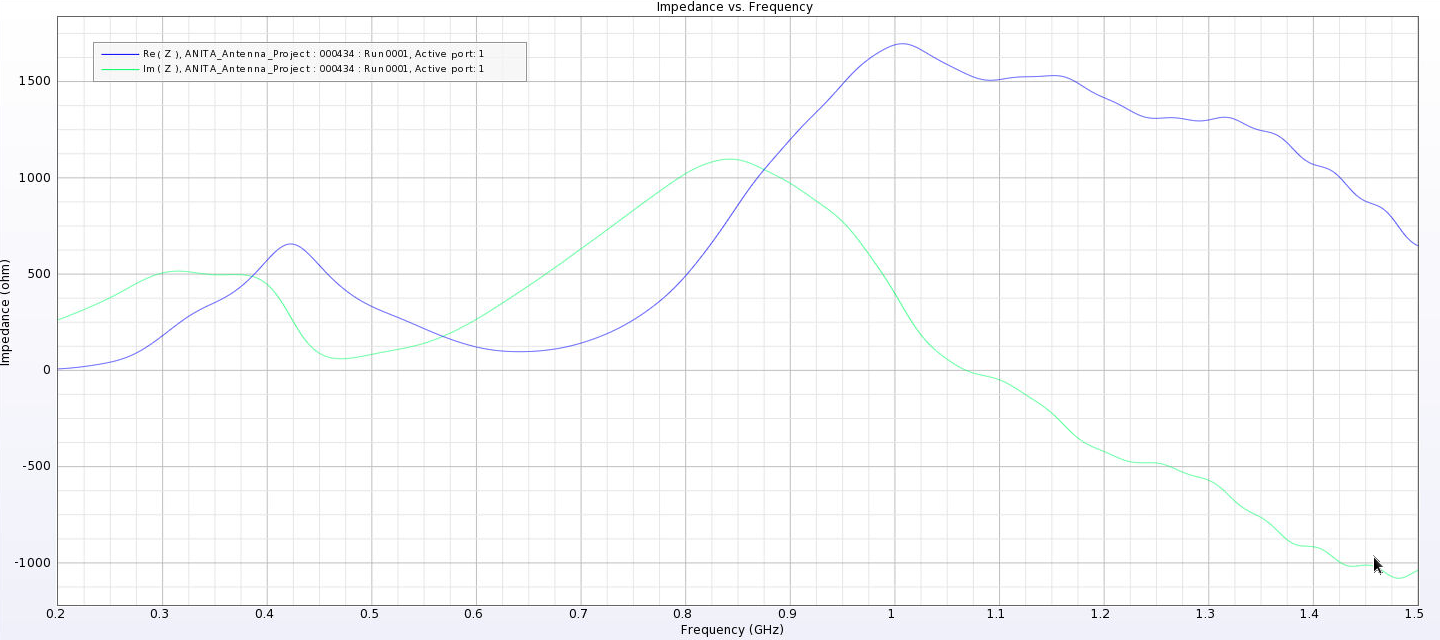 |
| Attachment 12: Separated_Impedance_24_cm.png
| 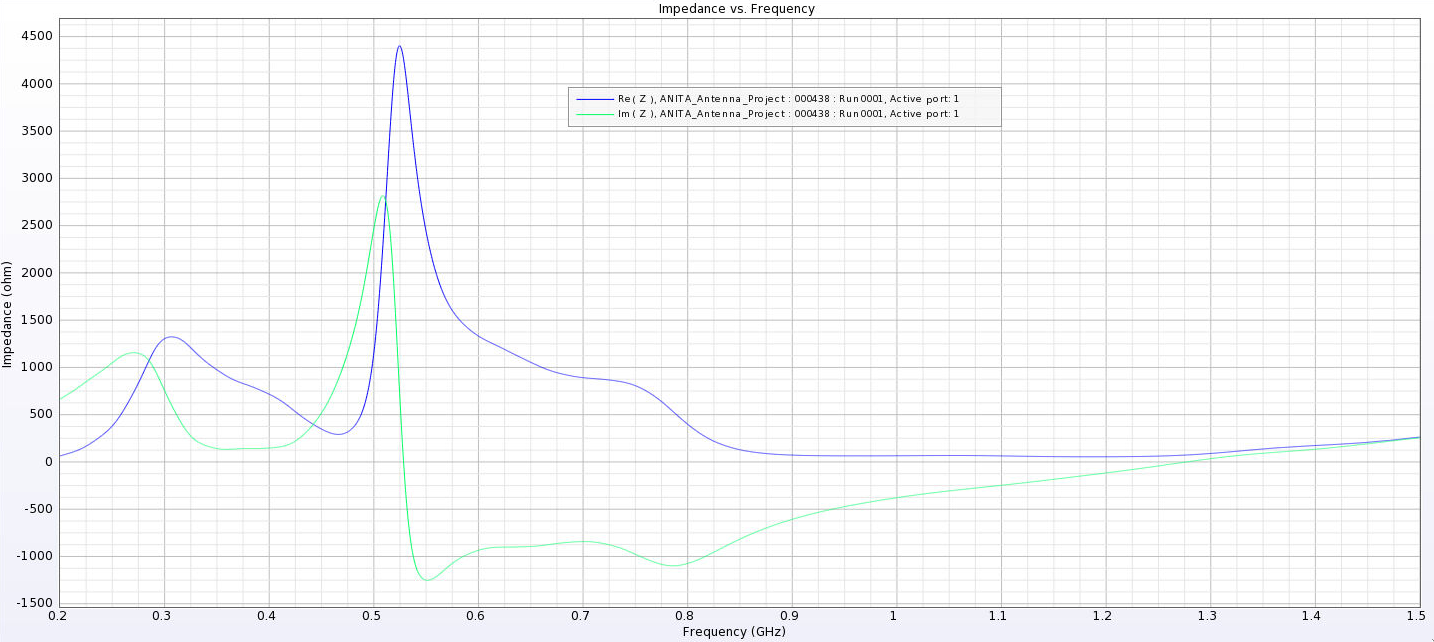 |
| Attachment 13: Separated_Waveguide_Impedance_1.png
| 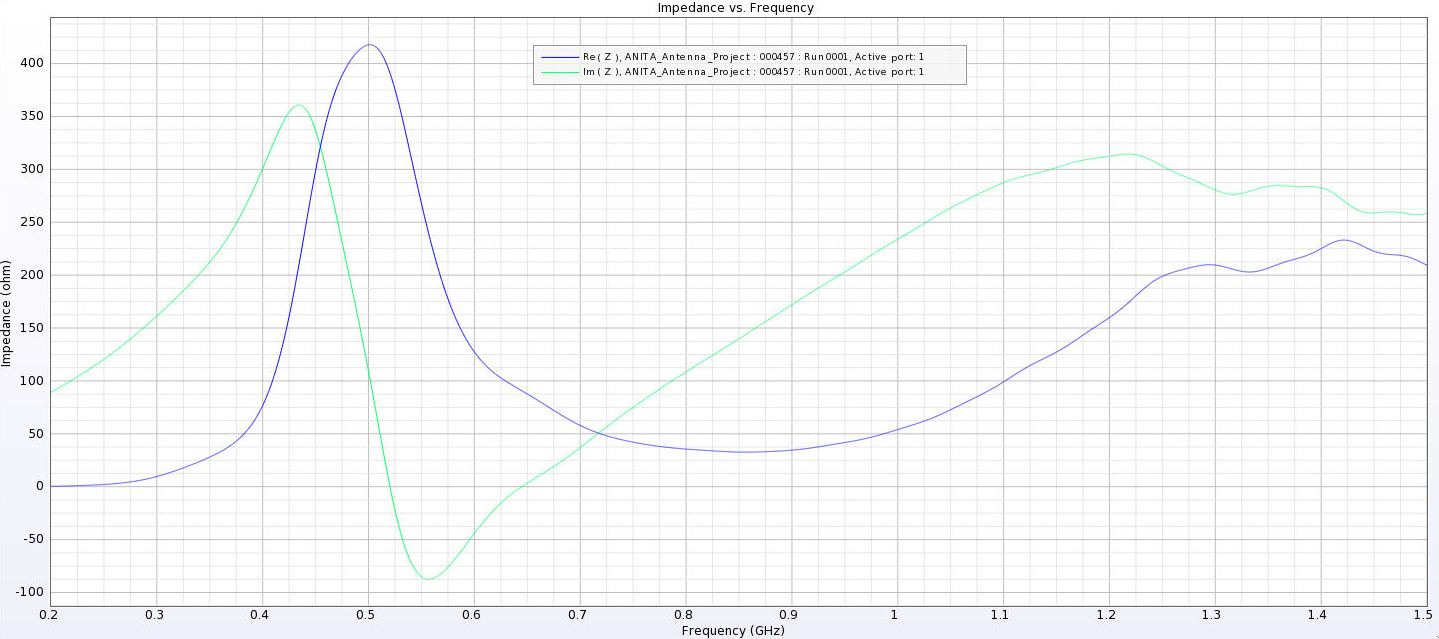 |
| Attachment 14: Separated_Waveguide_Impedance_3.png
| 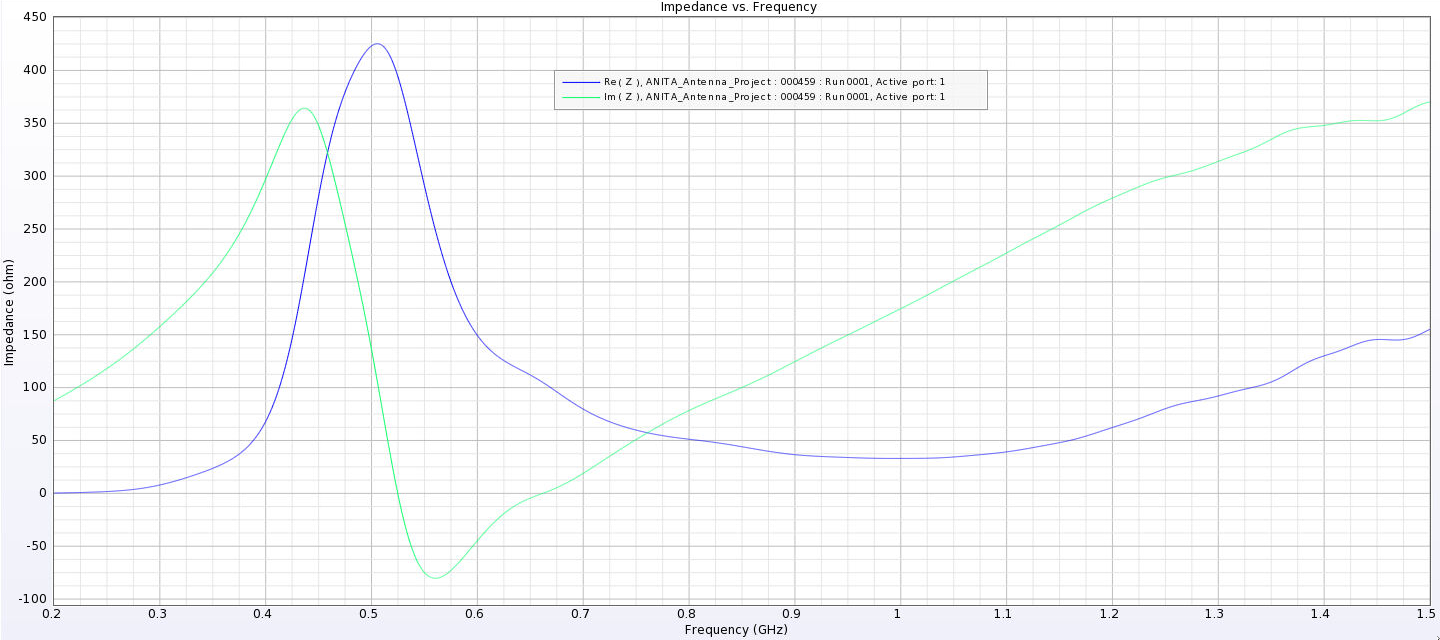 |
| Attachment 15: Separated_Waveguide_Impedance_6.png
| 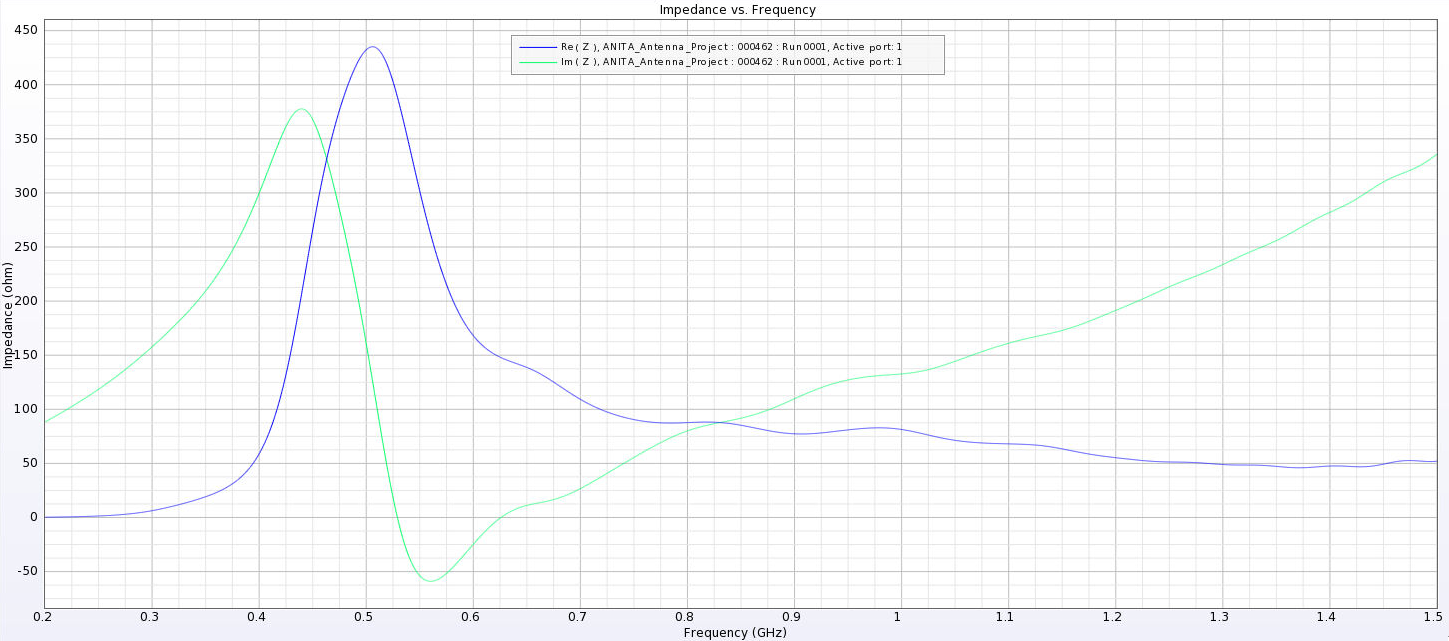 |
| Attachment 16: Separated_Waveguide_Impedance_8.png
| 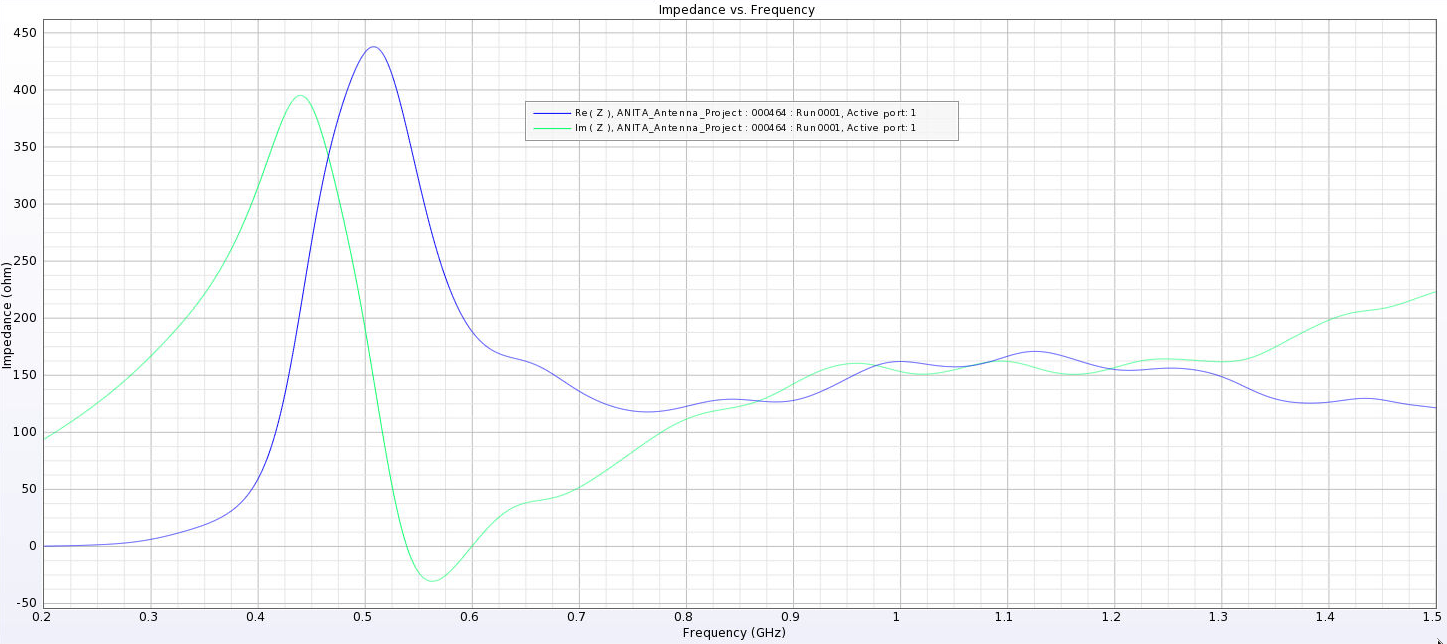 |
| Attachment 17: Best_Impedance_Plot_Horn_Antenna.png
| 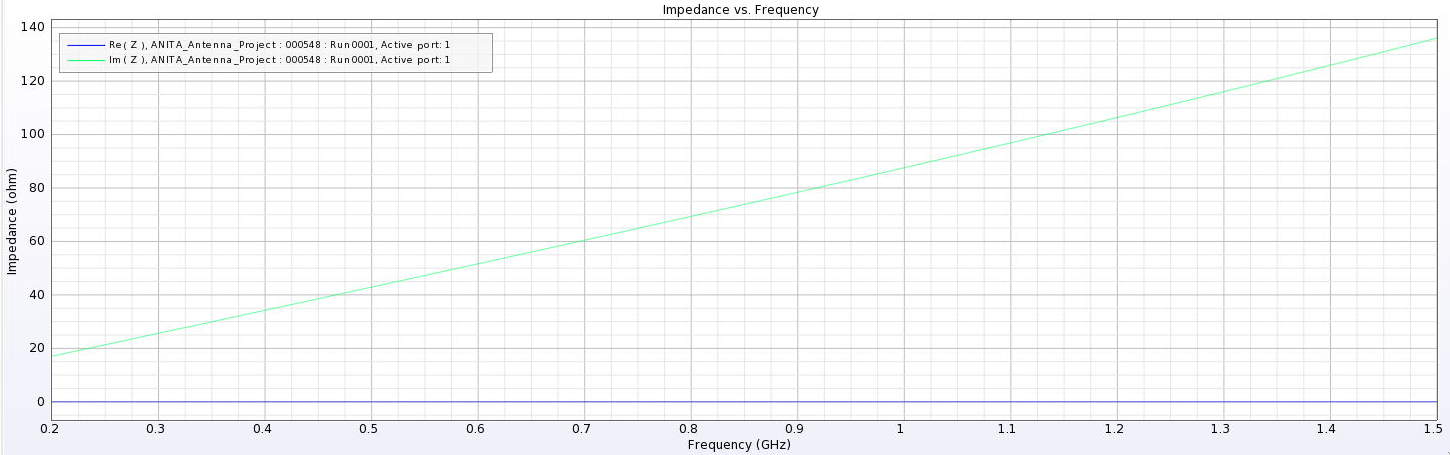 |
|
|
30
|
Tue Feb 4 17:17:10 2020 |
Julie Rolla | GENETIS working meeting | As of 01/30/2020, the proposal was submitted! The project description and summary are attached below.
Bicone Loop Status Update:
The loop currently runs; however, it does seem that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
The loop currently has the following additions implemented.
- Functionalized loop (XF_Loop_AraSeed.sh)
- Cleaner bash script which runs smaller bash scripts, instead of one massive script.
- Submits multiple AraSim jobs in parallel for each individual to increase speed.
- Ie if 100k neutrinos are thrown per individual ($NNT=100000, $Seed=10) we can, say, break it up into a "seed" of 10 -- which then submits 10 jobs with NNT=10k for each individual.
- Still needs this to be fixed: The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded. It is just submitting one job the following way.
- If NNT=100000 and Seed=10 (ie 10 jobs with 10k neutrinos), it will only run AraActualBicone as 1 run with 10k neutrino number (ie NNT/seed=NNT_New).
- Save State
- After every "part" -- ie part a, part b, etc -- of the loop, we save state by writing a series of numbers to a text file.
-
If the loop is killed for any reason during the process, the text file is read in, and the loop can continue forward from the last completed item/section.
-
This saves the state after every individual runs through XF.
-
Antenna sizing
-
The larger the antenna, the longer XF takes to run. A true-to-size ARA antenna takes anywhere from 1.5 to 2 hours per individual.
-
What we've done: Allowed us to scale the antenna by a factor of "x".
-
If we make the gaussian centered around something, say, 5 times smaller than the ARA antenna size, we then make the frequencies 5 times larger. This should have the same effect.
-
This also means we've had to scale these sizes back up when plotting.
-
Still needs this to be fixed: all of this has been hardcoded in! We need to make these variables in the MANY places we need to adjust for this scaling. See the to-do list below for details.
Moving forward, I will be doing the following:
- Getting our less coding-savvy teammates familiar with bash, python, and c++. This will be crucial for us when we start adding more parameters to our GA.
- Mastering XF (along with Alex, and Alex).
- Getting the team to clean up some messy coding done during the proposal hustle.
- Fixing some errors.
- Teach the ENTIRE team git.
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables.
-
Xmacros (use sed command)
-
All over bash script functions.
-
AraSim job files (sed command?)
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
|
| Attachment 1: project_description.pdf
|
| Attachment 2: project_summary.pdf
|
|
|
4
|
Wed Feb 6 17:36:24 2019 |
Julie Rolla | GENETIS update 2/6 | Atttendance: Julie, Max, Cade, Suren, Evelyn, Sophie
Today's tasks:
Max works on AraSim section (needs to do more research and reading)
Julie, Suren, Sophie, Evelyn work on plotting software to plot gain patterns of indiviudals
Cade preps impuse XF code to be implemented
To-Do list:
I. Divide up sections on paper, and work on them. See http://radiorm.physics.ohio-state.edu/elog/GENETIS/1 for section assignments.
II. Implement impuse code.
- This will require us to change XFintoARA.py so that it takes less output files.
- Is there anything else different with the output files?
III. Finish Evelyn and Sophie's plotting software.
- This means also puting it onto Nutau, and adding in lines to the bash script (both to move and save the images, and to run the code!)
Here are things we can do if we are waiting for Remcom:
I. Change sides of bicone to be sinusoid instead of lines.
II. Take known bicone parameters and put it through AraSim to check if it's working properly.
- This also means we should make plots with it to triple check.
III. Test GA and make it maximize other paramaters to see how it does. |
|
|
208
|
Wed Mar 29 18:36:29 2023 |
Alex M | GENETIS Working Meeting Update | This is an entry for the usual Wednesday working meeting GENETIS holds. We are largely carrying on from last week's hackathon hammering out the remaining pieces that need to be fixed. See the previous ELOG post for a list of those.
Alex
Amy pointed out on the Monday call that the proper way to simulate the ANITA antenas is to use one source at a time. I modified the script I had so that it makes two simulations per antenna. The first one will use the power source connected to the "VPol" ridges and the second one uses the one connected to the "HPol" ridges. After checking this, it looks like the asymmetry I had pointed out before fell away. In other words, I think XF only wanted to use a single power source anyway. So now we should be able to produce the VV, HH, VH, and HV data that icemc and PUEOsim need.
Ryan and I also worked a bit on the slopes of the ridges. We experimented with the parameter that affects the curvature, which seems to be able to be too large. But many of the designs still look to have almost no slope and many still caved inward rather than sloping outward. We're still working on figuring out what causes that, but I think it means we need another constraint on distance of the top of the inner ridges from the center.
We also tried figuring out why the sides of the curved part of the ridges don't stay in the same plane as the straight part of the ridges. I'm confident that they do stay in a single plane (I attached a Mathematica notebook that shows as much by finding that the torsion of the curve is zero; for some reason I couldn't print it to a PDF). We think that we should be able to rotate the curve into the same plane as the straight part of the ridge, but I haven't figured out how yet.
Nick and I continued working on the HPol antenna. We want to check if there are any other things in Steph's example XF file that are significant to the results of the simulation (versus just being there as a representation of the model). It looks like there are, and Nick is working to figure out what they are by deleting them (in a copy) and generating simulation results for them to compare to the original. Otherwise, the most significant parts (the copper plates) are now being made in a script and just need to be connected to one another.
Dylan
I tried to build root from source again and ran into an error 3 hours into the build. I messaged Will and asked if he knew anything about the error. I then reran the build with Will's suggested change and ran into the same error 3 hours later. I did, however, manage to build pueo by sourcing the installation of root in Will's shared pueo directory. I'm now testing getting a txt output file from the simulation as it turns out my initial solution didn't work.
Update:
I got a working version of root installed. Also, I was able to modify pueoSim to output a veff.txt file. Now, I need to modify where it reads in the gain files, and then modify the icemc specific bash scripts to work for pueosim. |
| Attachment 1: Modeling_ANITA_Ridges.nb
|
(* Content-type: application/vnd.wolfram.mathematica *)
(*** Wolfram Notebook File ***)
(* http://www.wolfram.com/nb *)
(* CreatedBy='Mathematica 12.0' *)
(*CacheID: 234*)
(* Internal cache information:
NotebookFileLineBreakTest
NotebookFileLineBreakTest
NotebookDataPosition[ 158, 7]
NotebookDataLength[ 141335, 3108]
NotebookOptionsPosition[ 137462, 3039]
NotebookOutlinePosition[ 137824, 3055]
CellTagsIndexPosition[ 137781, 3052]
WindowFrame->Normal*)
(* Beginning of Notebook Content *)
Notebook[{
Cell[BoxData[""], "Input",
CellChangeTimes->{{3.889104537246646*^9, 3.8891045516248693`*^9}, {
3.88910472825602*^9, 3.889104729415251*^9},
3.889118788593347*^9},ExpressionUUID->"f2d7bf67-519b-4366-9a10-\
f19376de6bb8"],
Cell["<\
Here, I will use an example ridge that I have made and tested in XF to \
demonstrate how the curved part of the ridges slope in.
Here is a figure that represents one of these ridges. We are viewing it \
directly at the edge of the ridge, so the blue line actually represents the \
side. The purple curve should lie directly aligned with the blue line but \
doesn[CloseCurlyQuote]t. The same goes for the green line and orange/red \
lines (which do lie in the same plane, but that plane isn[CloseCurlyQuote]t \
aligned with the other side) below, which is the other side of the ridge.\
\>", "Text",
CellChangeTimes->{{3.889118836397954*^9,
3.889119017191614*^9}},ExpressionUUID->"0859e4b4-741b-4eea-b2b6-\
e446b9af7816"],
Cell[BoxData[
RowBox[{
RowBox[{"x0", "=", "4"}], ";", " ",
RowBox[{"y0", "=", "4"}], ";",
RowBox[{"z0", "=", "0"}], ";", " ",
RowBox[{"yf", "=", "6"}], ";", " ",
RowBox[{"H", "=", "30"}], ";", " ",
RowBox[{"zf", "=", "30"}], ";", " ",
RowBox[{"xf", "=", "zf"}], ";", " ",
RowBox[{"[Beta]", "=", "2"}], ";",
RowBox[{"[Tau]", "=", "0.26"}], ";"}]], "Input",
CellChangeTimes->{{3.8891047313587914`*^9, 3.889104784350019*^9}, {
3.889105008478373*^9, 3.889105011515078*^9}, {3.889106004732277*^9,
3.88910602661027*^9}, {3.889112979624475*^9, 3.889112979661337*^9}, {
3.8891130240801353`*^9, 3.889113037799301*^9}},
CellLabel->
"In[433]:=",ExpressionUUID->"fb3dee39-fae9-47d5-bb54-52d21df6980e"],
Cell[CellGroupData[{
Cell[BoxData[
RowBox[{"ParametricPlot3D", "[",
RowBox[{
RowBox[{"{", "[IndentingNewLine]",
RowBox[{
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{"y0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "z0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
"}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{"-", "y0"}], "-",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "z0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
"}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"xf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",", "y0", ",", "z0"}], "}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"xf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{"-", "y0"}], ",", "z0"}], "}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"xf", "+",
RowBox[{
RowBox[{"xf", "/", "[Tau]"}], "*", "t"}]}], ",", "yf", ",", "zf"}],
"}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"xf", "+",
RowBox[{
RowBox[{"xf", "/", "[Tau]"}], "*", "t"}]}], ",",
RowBox[{"-", "yf"}], ",", "zf"}], "}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"xf", "+",
RowBox[{
RowBox[{"zf", "/", "[Tau]"}], "*", "t"}]}], ",",
RowBox[{"y0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{"zf", "/", "[Tau]"}], "*", "t"}]}], "}"}], ",",
"[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"xf", "+",
RowBox[{
RowBox[{"zf", "/", "[Tau]"}], "*", "t"}]}], ",",
RowBox[{
RowBox[{"-", "y0"}], "-",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{"zf", "/", "[Tau]"}], "*", "t"}]}], "}"}], ",",
"[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{"y0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{"[Beta]", "*",
RowBox[{"Log", "[",
RowBox[{
RowBox[{
RowBox[{"t", "/", "[Tau]"}], "*",
RowBox[{"(",
RowBox[{
RowBox[{"Exp", "[",
RowBox[{"zf", "/", "[Beta]"}], "]"}], "-", "1"}], ")"}]}], "+",
"1"}], "]"}]}]}], "}"}], ",", "[IndentingNewLine]",
RowBox[{"{",
RowBox[{
RowBox[{"x0", "+",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"zf", "-", "x0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{
RowBox[{"-", "y0"}], "-",
RowBox[{
RowBox[{
RowBox[{"(",
RowBox[{"yf", "-", "y0"}], ")"}], "/", "[Tau]"}], "*", "t"}]}],
",",
RowBox[{"[Beta]", "*",
RowBox[{"Log", "[",
RowBox[{
RowBox[{
RowBox[{"t", "/", "[Tau]"}], "*",
RowBox[{"(",
RowBox[{
RowBox[{"Exp", "[",
RowBox[{"zf", "/", "[Beta]"}], "]"}], "-", "1"}], ")"}]}], "+",
"1"}], "]"}]}]}], "}"}]}], "[IndentingNewLine]", "}"}], ",",
RowBox[{"{",
RowBox[{"t", ",", "0", ",", "[Tau]"}], "}"}]}], "]"}]], "Input",
CellChangeTimes->{{3.889104789299713*^9, 3.889105080982005*^9}, {
3.88910513362922*^9, 3.889105134787335*^9}, {3.889105176753415*^9,
3.889105183526766*^9}, {3.889105234070648*^9, 3.8891053106544733`*^9}, {
3.8891053894877863`*^9, 3.889105467143161*^9}, {3.889105814148295*^9,
3.889105946276293*^9}, 3.88911016079349*^9, 3.889111844551866*^9},
CellLabel->
"In[434]:=",ExpressionUUID->"a2747aaf-59c9-498b-a918-186b6556d2ae"],
Cell[BoxData[
Graphics3DBox[{{{}, {},
TagBox[
{RGBColor[0.368417, 0.506779, 0.709798], AbsoluteThickness[2],
Line3DBox[CompressedData["
1:eJwVjHk41Isexh1ahha/2fyQiIZkG2Mf/JrXcitCi4RzSUI8t1G3EUqbJCod
RyqpQ9f2KGSo64QkmpTUTWWP0mI5kVD2/c75432+z/t5Pu9Xc+/BHUGyMjIy
rtLISePx7o+1MjIEVK0qZf++4511NZyEnA0FF57G6NEINETP1GdIedNN5aN2
bTGCX2yHPu8kCFyo4/9h9wsBsF/vVW5LFohv2mfkrSEw/et90zOyBCK25bfK
tWUJlgT2qbnbEXh3oihwyWIC9EEyear1nqB00EIpN4ZA6pPMZ8sVCHx86HZw
tFUiUFnUQHNZTIfS1rgLKiwCGzn9H360Ngg+GNEGrykwQN+3lWGqS2Cq1fLq
cOsXgc6zO10cOxaCzj6S1LoTkDjcynmbNSKoD35xUmeHEmaXilYfPU5g1cfy
3r0Gc4KJNxkptV7KOC/apLE7hwBT6Je/1EEOn44VvfNNUMXh+qnHOa8JJPE2
Vrh50KDHs5SEpKoh87fhlbRJAq2pxtU/iBUw051oDnmrDv0NNbP/W0tHQs6p
17IqdPiNWJouil6DOy3ll5660UFe6Tp/OZ2JplZjt8RYTRxsGHnTHkFH/E6n
DQ3jbOh9eDrP+I8WkqoU7m7PoiOMn+lXF6MMcTAp25C7FqOK+2x6XtDhODQd
daBLFaMeT5/syePAdXOX8M5POmJst/urWq2GapDiqv7H2lgkogTNqxmwEV8v
OVmlgQNT7KeFtTqYm++oUN3IgHDyR/TzFZrIEZctEnatw84Tnu8eH2JgUp/o
NPTQwvXaUtnpKV1kdo2mp6UxYF1wJyMlZS0eBYW5S+bWg1fYsvxKDQNDDOc9
uyQc1HBe8s+S+lAPuar2fYiBLxf37BIt1cFUyEdRjLoB3OJSW/YqM7FpKNKp
x2IdiPrf7aJsDNFx9QjP2pGJbwG0sJJTuuhNSau3czfC7t+iTP9xkAlHDikb
L1mPge4gsYMXF5yv1R+F15ggXukkFg/poS+vh57BN8Zms26DcQkTq84VbbOw
MMBo9nh4RaUxxF1bOGnfmLimzyeYwYb49Nq8fIOAB4VjzZJwJgsvVO3MRy4Z
wXa5w6aqlzzUyNnK3t3AgrWaJInbwgU3QTz/3s0E1UO932jBLCjKZR32jzFG
supXmcn3JlB9dSBCksyCxmkNW28dHlo6tn6JCjHFv5KVbxdVsrBc2+f+jloe
/vuR/f1GvylmHyyJLe9hgUepFEX6mmBhJM++LNIMA01vZbTpbIR3Mp44y5ii
yzntsWjGDOo7Pq8r5LPxc+Sih1OqKW41PtS4etwcm0oZ4yH+bDhzn91wMjRD
h7DgeQnNAq9WaAbFXWRDMmIzM1FvhlubJ28cSrBASm3YyeoSNkQGJu1vg8zR
ZcYs01exxENRtpXPJzbsBwObvOUtUBZwcT4m3RLap8l0lrwSnDnKriuyLCCs
MeXYa1ghNrLwtgxPCUbRF45ILC2RZJ53Zb7ACvBb4+HmowTjIH6VdrslXPqO
G3lx+RDktxaUxSpB+CgktuSQFY4WqjOLKvlI692fHVushNqMpsAfbD7isgdb
Qp2sMaigZR/boYS52wsT3FI+9N6r1dS9tMaZZLWEDDkSQu0C+Ruu1hBoq8xq
edsgYtnoYXUuiRm9yxb3hq0xUBzmq/LBBgV/KcqXe5KQ5Mx8HjtngyMHZtMj
d9uipcDVMf4UiUDHtbBRt0W4rfv6xn5b+PmHrCnNJ6EdGl9uVG0LeVda58tQ
CnpRtblyjSQ0FIR3XT0pzP0uFLP+TWFvsGZqqZSLHU5EDXtTGGx4c9JXRCF1
57Hz+5tI2JxIdLjsQ6HR67rGUASFJVyusLGZhOdwcXObP4X0QD1/RjSFL10p
vOw2EoktY1MBQgq84y7dXskUrrvue2jfSWI+65R91GkKvxYk9ffcp0DTXNxw
uo9Ep83lxtwCCi6Dow+MyikIVvo9Me8nsT88J81JLO0874TICgoRM2UlfVI+
Lf4zaKCYgnappoF8NYXuZuG17QMklLXaJnjSv0OSe6EGdRSqLzT5aA6R2Epb
rVYp9c60Nw2LOihEjmb/VT1KoqopN7CxWfp3Nf9xxQcK4s9zbYfHSLitLDOM
aKMQsif90qJPFHrqPV/ojpPo2FQ3rizdu/YGmaR0U3DPW1aYNEFi4sG3c7ul
HvlzPKz8OwWj3WGiPdMkjDKNC/v6KSiY/9NRdpjCvi2vAlgzJCrb7SIuSr3Z
yCrWlp8U0qzWeTyXcmeWu4Ar9T7Pxf/5fly6Y3ZYGs+R2Bcf/vbwGIUmDJzV
nqJgJ2O+vlvKx6rjbpCTFJ6d2bbrwAyFI98TVVPnScRMXwt4ME2h9FmJTukc
haL2r8tcFkgomuUZ+Ep7vrzKxMIChd5a+7kFKb8Z+mDs7/5/SZ4y2A==
"]]},
Annotation[#, "Charting`Private`Tag$112685#1"]& ],
TagBox[
{RGBColor[0.880722, 0.611041, 0.142051], AbsoluteThickness[2],
Line3DBox[CompressedData["
1:eJwVjHk81Psexh1UQ4vfbH7IEs1ItjH2YX7Nx3IrQouEc0mWKa/bpNvYSpsk
CseRCjl0bS8lGdV1siXfJpG6qewRkuVEQtmX4c754/njeb/ez6MdcPIAX1ZG
RsZVGjlpPD7+sVVGBgM16xopxtBsb2MdI7FgR3HCyxh9EgbN0UtNOVLeekfl
jF1nDO8X7kT/QQyDhEbOH3a/YAjo7wJUOlN5ojv2OUVbMFj89YnZZVkMRey7
3yHXmcdbGzSi7m6HwcfzpUFr12CIPI6nLnQ85pWPWyoXxmCQ8SK3foMihvqe
up2c7hDzVOWbSS5ryKC8Ny5BlYahnYzRnh8dzbweY9J4uiIFyEf3Usz0MLTQ
YXVrsuMLT7f+wQDDjgb8K8/EDe4YEjvcLfiQN8VrOvb6gu4BZVheJ9Q4cw5D
m/sqhwMMJby59zlpDV4qcE24S+twAYaoAr/76xzk4PPZ0o++iWoQ1rTwvOAd
hlLYO6vdPEigz7YSB2eoQ+5vk5tI8xjqyDBBP7CNYK431xb8QRMMdtQt/28r
GSUWXHwnq0oGvykrM/noLfCgvfL6Szcywm8OXLuRTYXWDhO35FhtONk89b4r
goziDzrtaJ6lg37PyxXKf3QgpVbx0f48Mgrl5Po1xqiA6Bgu21y4FaaVjtoO
vSYjx4nFqJABNZj2ePniSBEDXHcPCB78JKMY7n5/NWsNUOMrbR59zgR5IcFr
06AgW9Htsgu1WhCyQH9Z0qALkpXuarWdFCSY/xH9aqM2FIgq5AUD2+Dgec+P
z09R0LwB1mvkoQO3G8plFxf0IHdgOjsri4Jsih/kpKVthWf8UHexZDuwS9o3
3KyjoAmK85FDYgbUMd5wruAGoBl8S/37BAV9STpySLhOFxaC+4QxmobgFpfR
HqBCRbsmIp2GLLcB1vS7XZStEXTfOs22caSib4Gk0LKLejCcltVk524Mh3+L
MvvHSSpyZOCy8eLtMDbIFzl4sYDxFfUJ0qkIe6ub/HBCH0aKhsg5HBPYbT5o
OCumos1XS/dZWhrCdP5seHWNCYgG9jCyvlFRugEHox4zgs/vLCp38NigeLZN
HE6loddqdhZT142Bu8FhV+0bNtTJcWUf7aAhG3VxCqudBaxE0conN1NAE8Pf
SMdoSEkuL8w/xgRS1b7KzH8yBbW3IRHiVBrSuqTF9dZlQ3v33i9RwWbwr1SV
e6U1NLSB6fPkQAMb/ttH/545agbLVWtjK4doiE2olkb6msLqVJF9RaQ5jLV+
kGGS6Si8l/LCWcYMBpyznguXzEHzQP+2Eg4d/ZxK8nDKMIO7LU+1bp2zgF3l
lNlgfzpyZtVnOhmZQ7eg+FUZyRLebtTmxyXRkXjKdmmuyRzu7p7PPJVoCWkN
oRdQGR0JDU27PvAtYMCcWmGgagVPhfnWPp/pyH48qNVbwRIqApNWYrKtgHkJ
z6YpKCNnhorrxjxLENSZMey1rCE2suSeDFsZGUcnnBZbWUGKRdHNlWJrAL8t
Hm4+ysiEz6lldlmBy8g5Yy8WB3j3O4orYpWR4FlwbNkpazhTokktreFA1vDx
/NiHyqghpzXoB50Dcfnj7SecbGBcUcc+tlsZSe6tzrHKOaD/Sb2u8Y0NXE5V
T8yRw5GAWayQ6WoDPKbqso63LUSsnw7TZOFoSf+G5eNJGxh7GOqr2mMLxX8p
KVR64khcsNQ/c9UWTocsZ0ce5kJ7satj/EUcBTluBVtNLoRz3be3jHLBzz94
... 2817 more lines ...
|
|
|
38
|
Wed Mar 4 01:00:35 2020 |
Julie Rolla | GENETIS Working Meeting 03/03/2020 | Attendance: Julie, Mitchell, Ryan, Alex P., Leo, Alex M.
Today's Tasks
Mitchell: Worked on Python Project and helped with Ryan's final bash project questions
Ryan: Finished the bash project, and is about to start the Python project.
Leo: Worked on lectures, which will restart on March 17th.
Julie, Alex P, Alex M: Worked on fixing the job XF submission scripts. Worked on making it so that it saves state after each individual's XF simulation is complete. We tried to parallelize as many of the XF scripts as possible, but if we have more than one it asks us to choose a license for each individual -- ie requires user input each time. Since we haven't found a way to get around it AND since running with 1GPU is super fast (under 5 min per individual), we are going to run them in series.
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF. (PASSED THIS TASK TO JULIE. WILL DO TOMORROW 3/4)
- Evolve: Right now, we are using an interactive job with only a CPU running at half scale, and have just edited our software so that we submit jobs for the XF simulations requesting 1GPU (could be 2GPU if we choose to edit it so).
- WHY WE ARE DOING THIS:
- Remember that we only need an interactive job when XF initializes and reads in the DNA for each individual. The GUI closes once the simulations start and we no longer need an interactive job.
- We were requesting a 1GPU interactive job and just running XF from the command line; however, OSC has been SUPER busy lately and we haven't been getting them (especially since we need long wall times if we are running the full simulations over many generations within the interactive job). However, we realized that if we submit an interactive job at a CPU (so that XF can allow the GUI to pop up), and then run the simulations by requesting 1-2GPUs we can then request a much smaller wall time and it will be quicker for the simulations to run --- ie we would be requesting a wall time of about 10 minutes using 1-2GPUs per simulation for each job submission versus 5+ hour wall time of an interactive job using 1-2GPUs.
- WE HAVE TESTED THIS, AND WE HAVE ERRORS. WE WORKED ON FIXING THEM TODAY, AND HOPE TO FINALIZE THEM TOMORROW (3/4) AND RUN!
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
As of 3/3, I have not gotten an update on this. I will check on the status of this tomorrow.
-
Comment code thoroughly (Everyone needs to help with this)
-
Julie made some progress on this over the weekend.
-
Made comments on what each bash script version does, and cleaned up our directories from old code so that it doesn't confuse the others when they start helping
-
WAY more needs to still be done.
-
Get new proposal results for March 12th deadline.
|
|
|
31
|
Mon Feb 10 14:20:56 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Write bash practice assignment, assist others when they have questions, work on Git lecture. (Will finish bash assignments early next week)
Evelyn, Mitchell, Eliot: Make progress on the "Learn the Command Line" course on Code Academy. (Should be done by the end of the week?)
Leo: Start gathering information for the "physics of antennas" lecture series! Note: Remember that Leo wants to be an educator, and would like us to find opportunities to work on "teaching" items. Most of the students don't have any background on how antennas work. He started learning about them and is going to do a lecture series (likely the first 20 minutes of every meeting) starting in a few weeks! (Should start series by hopefully 2/21)
Alex M: Clean up unused code. Fix hard-coded directories (likely finished & tested by end of day 2/11)
Alex P: Fix hard-coded variables (likely finished & tested by end of day 2/11)
I've tacked on our to-do list in the meantime so we can keep carrying the items still pending along with us. As we complete them, I will drop them off the list!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables.
-
Xmacros (use sed command)
-
All over bash script functions.
-
AraSim job files (sed command?)
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
|
|
|
32
|
Wed Feb 12 01:18:22 2020 |
Julie Rolla | GENETIS Working Meeting | Today we continued with the to-do lists items previously posted. I marked all the to-do list items that have been completed with green and a slash. Note that I have also added a new item at the bottom of the to-do list.
Today's Tasks
Julie: Write bash practice assignment (almost done!)
Evelyn, Mitchell, Eliot, Ryan: Make progress on the "Learn the Command Line" course on Code Academy. (Mitchell is done, and the others are close.)
Leo: Start gathering information for the "physics of antennas" lecture series! Note: Remember that Leo wants to be an educator, and would like us to find opportunities to work on "teaching" items. Most of the students don't have any background on how antennas work. He started learning about them and is going to do a lecture series
Alex M: Make sure antenna scaling is made a variable in the bash script everywhere that it exists, start mass commenting all of the code so it reads as cleanly as the GA does. (Finished making the antenna scaling a variable. needs to be tested, but we waited forever for wall time)
Alex P: Fix hard-coded variables (done but tried to test it. Waited for wall time and never got it), and looked into ways to get around requesting an interactive job. For some reason, we don't need an interactive job to forward things like plots, but we do for the XF UI. We were trying to figure out why this is, and if there's a way to fix this. So far, no luck.
AMY QUESION: Could we actually consider running on NASA computers? I think Alex P. found a way the NASA computing centers let you adjust for this issue, and it didn't work on OSC. If we end up not finding a solution with OSC, is it at all possible to have XF installed with NASA computing centers? (NOTE: I JUST ASKED ALEX FOR MORE DETAILS AND WILL ADD THEM HERE TOMORROW -- IE WEDNESDAY)
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim error fixed (Alex, Julie)
-
It seems that if AraSim jobs are left to run while the user is away, it doesn't properly write one or more of the flags, and makes the loop get stuck at AraSim. We must then re-run from that state by manually editing the Save State text file.
-
We are going to try to get XF to run at the command line so that we can submit it as a job. This would alleviate us using an interactive job. We *hope* this would fix the error since the user wouldn't be "gone" while utilizing a GPU in an interactive job.
-
Alex emailed XF to ask a few questions! Stay tuned!
-
Make Hardcoded directories variables
-
Make the following variables in the bash script:
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
In roulette_algirithm.cpp
Length = 50cm/Multiplier_factor;
STD= 15cm/Multiplier_factor
Radius = 1.5cm/Multiplier_factor;
STD= 0.75cm/Multiplier_factor
Also the "Multiplier_factor" line 89.
This "Multiplier_factor"has to be the same as the GeoFactor in fitnessFunction_ARA_amy.cpp AND the same as the factor higher in frequency in the xmacro.
NOTE:THIS MUST BE RECOMPILED AFTER CHANGING.
In fitnessFunction_ARA_amy.cpp:
GeoFactor=2
This GeoFactor has to be the same as the Multiplier_factor in the roulette algorithm AND the same as the factor higher in frequency in the xmacro
NOTE: THIS MUST BE RECOMPILED AFTER CHANGING.
-
Get rid of all code we are not using anymore.
-
Currently using: XF_loop_AraSeed.sh
-
Look at what items are being called in here, and remove old versions such as XF_loop_Prototype.sh, XF_loop_Functionized.sh, and the related scripts.
-
These can temporarily moved to on "old code" directory and we can delete once we finalize the loop.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being seeded.
-
Comment code thoroughly
-
The Gen alg is commented so thorougly that it can read like a book. We want this with ALL of our code; it will help all the less familiar people get comfortable.
|
|
|
33
|
Fri Feb 14 16:44:44 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Write Python project! I have officially finished the bash course for the students. It can be found here: https://docs.google.com/document/d/1nGdPrwfYJOrO6NBT76Wsh6T0hDmGHo2-23oJ1WyeNcs/edit?usp=sharing. Mitchell is currently trying it out and will give me comments.
Mitchell: Work through my bash project and give me comments!
Evelyn, Eliot, Ryan: Make progress on the "Learn the Command Line" course on Code Academy.
Leo: continuing gathering information for the "physics of antennas" lecture series! These will take place every Friday meeting starting next week.
Alex M: Try to get scaling into the xmacro, comment the code better.
Alex P: Add on the AraSim fixes mentioned below!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
Carl believes this is due to the way we are saving our AraSim outputs. Instead of saving the files directly to our desired location, we are going to save them to $TMPDIR and use "pbsdcp" to copy them from $TMPDIR to our desired location. This may alleviate the problem since it will not be writing all the files to our desired location at once.
DATA_LIKE_OUTPUT=1,2 doesn't work with DETECTOR=0,1,2
DATA_LIKE_OUTPUT controls data-like output into UsefulAtriStationEvent format; without a real station selected (using DETECTOR==3,4), the mapping to the data-like output will not function correctly
There are 1 errors from settings. Check error messages.
SysError in <TFile::Flush>: error flushing file outputs/AraOut.setup.txt.run29.root (Stale file handle)
- Explanation of issue (2): We keep timing out of the interactive job when AraSim is running (if the user isn't remaining active on their terminal).
- OSC suggested that we use a virtual desktop (via ondemand) insead of using an interactive job. We will try this.
- Note: We should now be requesting 2GPUs instead of 1GPU.
-
Make the following variables in the bash script.
-
Alex M is working on this. We can't directly make this a variable. We need to take the array of frequencies and multiply it by some value and THEN feed it into the xmacro. He is looking into whether of not he can directly do this in bash. If not, I have written a small python script that does this, and he can write it to that file; however, he can't append it at the end so it's not the ideal way. We are going to continue to try to do it through bash first.
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
The Gen alg is commented so thorougly that it can read like a book. We want this with ALL of our code; it will help all the less familiar people get comfortable.
|
|
|
34
|
Tue Feb 18 16:20:59 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: Continue writing Python project!
Mitchell, Evelyn, Eliot (not here on Tuesdays), Ryan: Work through bash project
Leo: continuing gathering information for the "physics of antennas" lecture series! We start this Friday at 4pm.
Alex M: Try to get scaling into the xmacro, comment the code better.
Alex P: Add on the AraSim fixes mentioned below! He is testing this now. We are waiting for updates.
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
UPDATE: Alex P. has edited our bash job script to save the outputs to $TMPDIR and then to pbscp (submit a "job" to copy them over) them into our correct directory that the bash script needs them in. He is testing this now and will report back.
-
Note: we still need to make sure that every time we run that we divide AraSim runs into MANY more jobs (ie further parallelize by dividing into, say, 100 instead of 10).
DATA_LIKE_OUTPUT=1,2 doesn't work with DETECTOR=0,1,2
DATA_LIKE_OUTPUT controls data-like output into UsefulAtriStationEvent format; without a real station selected (using DETECTOR==3,4), the mapping to the data-like output will not function correctly
There are 1 errors from settings. Check error messages.
SysError in <TFile::Flush>: error flushing file outputs/AraOut.setup.txt.run29.root (Stale file handle)
- Explanation of issue (2): We keep timing out of the interactive job when AraSim is running (if the user isn't remaining active on their terminal).
- OSC suggested that we use a virtual desktop (via ondemand) instead of using an interactive job. We will try this.
- Note: We should now be requesting 2GPUs instead of 1GPU. UPDATE: Alex P. requested one, but it took too long and he had to leave before he got it.
-
Make the following variables in the bash script.
-
UPDATE: Alex M. figured out how to pass the scale factor into the xmacros. We used the "tr" command in bash. He will be testing this and can update us at the Thursday meeting.
In xmacros/simulationPECmacroskeleton_prototype.txt (use sed command):
Grid spacing=0.1
Frequency= factor of n higher
This factor of "n" higher in frequency has to be the same as the Multiplier_factor in the roulette algorithm AND the same as GeoFactor in fitnessFunction_ARA_amy.cpp.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
UPDATE: Alex commented code that utilized the scale factor (Part_E.sh). More still needs to be done.
|
|
|
35
|
Fri Feb 21 16:47:21 2020 |
Julie Rolla | GENETIS Working Meeting | Today's Tasks
Julie: I finished the Python assignment. I am making sure my solutions encompass all of the different ways to solve this, and will then start on the C++ project.
Mitchell, Evelyn, Eliot, Ryan: Work through my bash project.
Leo: At 4 pm today Leo will begin his "physics of antennas" lecture series!
Alex M: All scaling is corrected to be set at the bash script level. Alex is now going to work on commenting code!
Alex P: Still working on our AraSim edits!
TO-DO List
The following is our coding to-do list:
-
Get our Ara sim errors fixed
-
Explanation of issue (1): Usually 1 of ~100 AraSim jobs never completes. OSC shows "completed", but the flag is never written and the output file is not filled in entirely. The error file shows the following.
-
UPDATE: Alex still trying to figure out how to input Car's suggestion. He also needs to add this for the flag files that are being saved. Here is his most recent question:
Alright so I have pbsdcp working with generic files and I now have it working in the AraSim call but my current worry is that it wasn't creating a file unless I had
./AraSim setup.txt $runNum outputs/ a_${num}.txt > $AraSimDir/outputs/a_${num}.txt
So I just did this to have some a_1.txt, before we had the last line writing directly to our antennaPerformance metric (this makes the AraOut file typically I just kept it as this name for testing purporses), now if I get rid of the portion after the > or the part after outputs/ it doesn't seem to make a file that we need. I think I don't understand to get the full output of AraSim without using this. Because I don't understand how using this is and then using pbsdcp is any more efficient.
Now maybe if I had the portion right after the > go directly into TMPDIR instead of using pbsdcp to both scatter to and gather from TMPDIR that would be better
-
Make the following variables in the bash script.
-
UPDATE: Alex M. finished editing the scaling factor to be at the bash level for every piece of code that uses it.
-
Turn tournament on in the GA.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized.
-
Comment code thoroughly
-
UPDATE: Alex commented code that utilized the scale factor (Part_E.sh). He is now going to begin commenting the other code to clean it up. We want it to be read as cleanly as our GA.
|
|
|
37
|
Sat Feb 29 02:21:16 2020 |
Julie Rolla | GENETIS Working Meeting | Attendance: Julie, Mitchell, Ryan, Evelyn, Eliot, Alex M.
Today's Tasks
Julie: Worked on C++ project, and helping others with the Python project; I also made additions, clarifications, and edits to the Bash project (also updated the solutions and pushed them to gitHub). I still need to do the following:
- Add the instructions from the google doc to the GitHub pages for both the Python and the Bash projects.
- Work on the C++ project!
- Run a test evolution this weekend to make sure AraSim stuff is fixed and that XF job submissions work.
- EVOLVE
Mitchell: Started Python Project.
Evelyn, Eliot, Ryan: Work through my bash project.
Leo: Worked on lectures.
Alex M: Helped make XF job scripts. Needs to do the following:
- Parallelize the AraActualBicone properly.
- EVOLVE!
Alex P: Needs to do the following:
- Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF.
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF. (ALEX P)
- Evolve: Right now, we are using an interactive job with only a CPU running at half scale, and have just edited our software so that we submit jobs for the XF simulations requesting 1GPU (could be 2GPU if we choose to edit it so).
- WHY WE ARE DOING THIS:
- Remember that we only need an interactive job when XF initializes and reads in the DNA for each individual. The GUI closes once the simulations start and we no longer need an interactive job.
- We were requesting a 1GPU interactive job and just running XF from the command line; however, OSC has been SUPER busy lately and we haven't been getting them (especially since we need long wall times if we are running the full simulations over many generations within the interactive job). However, we realized that if we submit an interactive job at a CPU (so that XF can allow the GUI to pop up), and then run the simulations by requesting 1-2GPUs we can then request a much smaller wall time and it will be quicker for the simulations to run --- ie we would be requesting a wall time of about 10 minutes using 1-2GPUs per simulation for each job submission versus 5+ hour wall time of an interactive job using 1-2GPUs.
- WE HAVE NOT TESTED OUR .SH JOB SUBMISSION SCRIPTS! JULIE WILL DO IT THIS WEEKEND!
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
Comment code thoroughly (Everyone needs to help with this)
-
Get new proposal results for March 12th deadline.
|
|
|
39
|
Fri Mar 6 15:01:37 2020 |
Julie Rolla | GENETIS Working Meeting | Attendance: Julie, Evelyn, Mitchell, Ryan, Alex M.
Since we have been having issues with the lack of availability of GPUs on OSC, we have come up with a new to-do list:
- Try to keep running on Pitzer with 1 core instead of 40. Based on first attempts, it is saying this will reduce wait times to ~3 hours (will vary based on usage), and after that 3 hours, human interaction will be needed. So the plan is, someone requests a job in the morning, and when the interactive job starts, the person is responsible for doing the thing which will allow the generation to run. We will aim for one generation per day under this plan, taking turns. Better than no evolving.
- I have asked Heechang at OSC if we can install the latest version of XF on Ruby and/or Owens. That would be better. There seems to be more available on Owens at least.
- Our initial attempt to run on Ruby is giving us an error which is probably to do with different inputs required in different versions. Will attempt to pursue this as a last resort, but the older version of XF does require lots more human interaction so until other options are exhausted it's not worth it.
Today's Tasks
Mitchell: Worked on Python Project, learned to evolve (note: Mitchell may leave the group at the end of the semester)
Ryan: Starting the Python project
Evelyn: Finish bash project.
Julie: C++ project, look at Amy's proposal draft.
Alex M: Fix XF solver to submit each individual as a separate job (thus we can request a smaller wall time for each job)
TO-DO List
- Scaling test run: Do a test run to see if the scaling (when we also scale the grid spacing) actually affects the run time for XF.
- We are going to pause on working on this, because we are struggling to get GPU time. We will test this whenever we see OSC less busy (ie if we log on at a lucky time).
- Evolve: Try to keep running on Pitzer with 1 core instead of 40. Based on first attempts, it is saying this will reduce wait times to ~3 hours (will vary based on usage), and after that 3 hours, human interaction will be needed. So the plan is, someone requests a job in the morning, and when the interactive job starts, the person is responsible for doing the thing which will allow the generation to run. We will aim for one generation per day under this plan, taking turns. Better than no evolving.
-
Parallelize the AraActualBicone job.
-
The AraSim run in Gen 0 that gets Veff for the ARA actual bicone input file is not currently being parallelized. (ALEX M)
-
As of 3/5, I have not gotten an update on this. I will check on the status of this.
-
Comment code thoroughly (Everyone needs to help with this)
-
Julie made some progress on this over the weekend.
-
Made comments on what each bash script version does, and cleaned up our directories from old code so that it doesn't confuse the others when they start helping
-
WAY more needs to still be done.
-
Get new proposal results for March 12th deadline:
-
Julie is looking at what Amy wrote here
|
|
|
108
|
Mon Sep 14 17:23:26 2020 |
Alex M | GENETIS Update 9/14/20 | I just wanted to give an update on what I've been doing today for GENETIS. I went ahead and recorded myself running the loop. It should be useful for new people we're onboarding but I also went through a whole generation, which I think is interesting for everyone to understand the timescale of each part of the loop. I'm making a small edit (I have no clue how to edit videos haha) so once I'm done I'll try to send it via email and post it on slack. I also started a long run Amy requested using antennas which are asymmetric in length and opening angle and 50 individuals per generation. Since it has so many individuals it will take a long time to go through each generation. I also had to decrease the number of AraSim jobs (thus increasing the number of neutrinos per job) because otherwise we'd be submitting too many jobs and have a lot of them blocked, holding us back from running. If anyone has problems with me using up too many jobs let me know and I can try to decrease the numer of jobs per individual futher (right now I have 10 jobs per individual with 3000 neutrinos per job, which gives us 500 AraSim jobs per generation). |
|
|
40
|
Tue Mar 17 16:18:29 2020 |
Julie Rolla | GENETIS Planning Meeting | Attendance: Julie Rolla, Alex M.
Today Alex and I met to discuss how to move forward given the COVID-19 restrictions. We have resent out a message polling people's schedules to create online working sessions. Here is what we are planning:
| Group name |
Tasks |
Participants |
Times per week to meet |
| Loop Group |
Running the loop, updating software, getting plots |
Julie, Alex M., Alex P. |
2 |
| Lecture |
Attend Leo's lectures |
Full group |
1 |
| Training (Python, c++, and Bash projects) |
Completing the training modules (Alex M. and Julie will be answering questions) |
Mitchell, Ryan, Evelyn, Eliot, Julie, Alex M. |
2 |
Once we collect times for each group, we will have separate meetings for all of these entities. This way it's easier for us to meet without all having different items to discuss/work on. All of these will be zoom meetings moving forward. Meeting info will be announced shortly!
Tasks for "the Loop Group":
- Make wall time a variable for XF sim.
- Test current software edits.
- Such as item #1 above
- Make sure the bottleneck function is working.
- Right now it seems to be working; however, we keep hitting a wall time error. If we can make the wall time a variable then we can be sure the bottleneck function is working, and the wall time is truly the error.
- Get Ruby working on XF
- when I type:
/usr/local/xfdtd/7.8.1.4/remcom/bin/xfdtd I get:
Info: Using latest available version (7.8.1.4)
Info: Running /usr/local/xfdtd/7.8.1.4/remcom/XFdtd_7.8.1.4/bin/xfdtd
Error: Unable to find executable to run. This command must exist
- Get Alex's account of Ruby working.
- He received an email and is still unable to log in. He is touching base with them again.
Tasks for "the Training Group":
- Julie and Alex need to finish the C++ project!
|
|
|
1
|
Wed Feb 6 17:23:18 2019 |
Julie Rolla | GENETIS Paper working draft | The current working draft of the GENETIS paper can be seen here: https://www.overleaf.com/6783528497tvqbjphsgvzn
Additionally, here's our outline for the paper. The section assignments are as follows:
I. Intro (Julie)
II. Types of Algorithms (Suren)
III. XFdtd (Cade)
IV. AraSim -- will be called something else... (Max)
V. Fitness function (Suren)
VI. Results (Julie)
VII. Conclusion (We will see..)
Note that the order of the sections may change. |
| Attachment 1: IMG_2881.JPG.jpeg
|  |
|
|
43
|
Thu Apr 2 16:14:29 2020 |
Julie Rolla | GENETIS Meeting update | This is a general update. Most of our recent meetings have been to teach the students to use Git properly, or to answer questions on the training modules.
Update
1. Git Update: I recently cleaned up our Git and GitHub so that we have a better way to develop this code. Once I cleaned it up the goal is to educate all of the students to properly use Git so we develop code correctly moving forward. The repository had become quite a mess. The students were constantly developing on a branch, and never merging it back to the main branch. This means that we never had a working copy that we could run on while edits were being made in parallel. I attempted to merge the edits to the main branch with the hopes of getting this functioning so we could evolve while working on the bottleneck function; however, it looks like someone with poor knowledge of Git edited the main branch and we had an abundance of merge conflicts.
What I've done now is reset the repository so that all merge conflicts are resolved and a working copy exists on the main branch. I've also added a .gitignore that will ignore all data files. Git doesn't like these because (1) it hates when tracked files disappear -- ie when we delete trial run data, if someone accidentally pushed it then Git complains-- and (2) it means it we constantly are adding more untracked files (every single new file we make is untracked and then gets tracked, like the flag files). So, I've had it only track useful code.
The goal once I train the other on Git is to move the place of the data files and make it it's own git repository (once we start doing fewer trial runs so we won't lose important data). If the others learn to check the status of files before pushing them, they can make sure they don't push useless trial data sets, and either (1) delete them, or (2) add them to the .gitignore.
As for now, we successfully have the working, run-able copy on the main branch now. When any more revisions are made (e.g. the talk of adding the wall time variable), we can branch and edit on that so we still run.
2. Evolution/bottleneck update: We got the loop working to the point that it can run all the way through a generation without fatal problems. The only potential problem we can see right now is if we need to go backward in the savestate. We're working on making that not a problem--the solution is to better organize the output files and data, which we were interested in anyway. Right now we are running with 10 individuals in each generation. Alex got through 2 generations without a problem, so he'll keep running but now we're trying to make some of these more minor fixes so we may have to stop the run. Alex does have plots (they don't look great, but at least they're being made correctly without error). At the moment we are swimming in GPUs. We haven't had much trouble at all this week with getting them, so that's why we were able to get 2 generations done between around midnight last night and 2ish today; not sure how long that will persist, but Pitzer has had relatively little usage whenever we've looked lately.
Comments on the plots: We're discussing this now, we don't think the individuals should really be color coded/separate on the legend because it is not like they follow one individual across multiple generations; however, we do need to label the dotted line. Another thing we were considering is decreasing the penalty factor for when the cones are too big for the hole. If we get something with a much much better Veff but it's only slightly larger than the hole, it might be worth it to help that one stick around a little bit more.
3. Wall time Variable Update: Also, last week Carl and Alex talked about the variable wall time again. We decided the best idea is to start by implementing a variable wall time based on the cylindrical volume that he mentioned before, but in the future, we might also be able to check the time of each GPU job and then plot it against the dimensions of the cone/cylindrical volume to get a better estimate of the wall time for each job
|
| Attachment 1: Fitness_Score.png
| 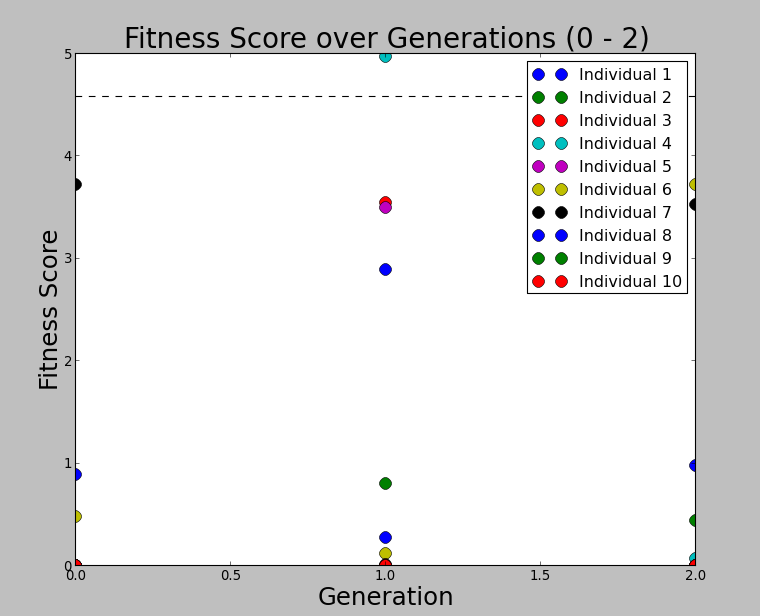 |
|
|
44
|
Tue Apr 7 15:48:54 2020 |
Julie Rolla | GENETIS Meeting Update | Meeting Attendance: Julie, Alex M., Alex P, Eliot, Leo, Evelyn, Mitchell, Ryan.
Today is the deadline for confirming our attendance at the virtual APS meeting. Alex M. will be confirming and we will be orienting ourselves so that we can have some results. Here is our to-do list for today:
- Create a method for automatically adding our plots to dropbox each day.
- The way we are going to do it is by adding the command: mail -s "Test Subject" someemailaddress@gmail.com < testScript
- We will create a random email to use for this that will only be receiving emails from this command.
- We are trying to find and set up a client that as soon as this random email address receives an email, it uploads it to the Dropbox here.
- This is still being worked on.
- Start developing a database of old antenna runs and parameters, and pull data from that for any repeat dimensions.
- This should speed up some of our run time.
- We want to create a database of old antenna dimensions and their associated UAN files. That way, if we repeat an antenna geometry very closely, instead of re-submitting a job to produce the gain patterns, we can pull from one that has already been created.
- This should limit:
- The number of jobs we have to submit.
- The number of GPU hours/minutes we have to request and wait for.
- These should probably still be run through AraSim, however, since the energy could change from run to run.
- Keep running and collecting data.
- Do we want to run and have data for 20ish generations with 10 individuals for APS?
- We have already made some progress on this so we could likely make more generations for APS if we keep going.
- Do we want to instead switch to doing something like 20 individuals even if we get fewer generations completed?
Notes apropos developing our database for antenna info. This is how we are going to do it, and what we need to develop in our code:
- Add all info to csv files.
- The csv files will have columns: length, radius, theta, uan file directory and name (ie /file/location/database#/).
- # will be the row that this matches the L, R, Theta within the csv file
- Inside /file/location/database#/ will be all the uan files for each frequency for these dimensions.
- We need an "if/else" statement in the bash script that tells the code to look in this csv file (read it in).
- IF the produced L, R, Theta match any of those that currently exist, save the 4th column (file location) to a variable (say we call it ExistingUan=/file/location/database#/).
- Then cd ExistingUan
- Make a loop that:
- does CP /file/location/database#/gen_individual_freq.uan BiconeEvolutionOSC/BiconeEvolution/current_antenna_evolution/XF_Loop/Evolutionary_Loop/Run_Outputs/$RunName/ (ie to the file name and location that XFintoAra.py reads. This will make sure that it has the data as if we actually completed the XF simulations for the antennas that had repeat sizes from our past runs (I mean past by runs we may have EVER done -- not past generations).
- We need to copy ALL of the frequency uan files for that individual into Run_Outputs/$RunName
- Else fill another line in that csv file with the L, R, Theta of this individual so we can keep it for later.
- mkdir the new directory /file/location/database#/
- Add the name of this new directory (ie database#) to the 4th column on the csv file next to the correlating L, R, Theta.
- Then submit the XF job for this individual.
- Once it finished the XFSolver job and produces the .uan files (for each frequency), do a loop that:
- cp individual_freq.uan /file/location/database#/gen_individual_freq.uan
- Note that I wrote to cp individual_freq.uan instead of gen_individual_freq.uan. Note that XF produces it as individual_freq.uan, and it is changed to gen_individual_freq.uan when we move it into the Run_Outputs/$RunName directory.
- So you can do it either (1) after it moves it to Run_Outputs/$RunName with the name gen_individual_freq.uan, or (2) before it moves the file with the name individual_freq.uan.
- We need to make a new directory that holds all of these database# directories and their uan files, and the csv file the "if/else" statement checks.
- We need to add this new directory to the .gitignore!
|
|
|
45
|
Mon Apr 13 14:43:54 2020 |
Julie Rolla | GENETIS Meeting Update | Note that Pitzer usage over the weekend was much better! Maybe weekends are our best time to run? Today (Monday) has been a little bit worse.
Over the past few days the following items have been accomplished:
- We got the jobs to stop canceling on OSC.
- Issue was the destination command for the error and output files for the GPU jobs inside the GPU job script; we fixed it by taking it out.
- This issue was causing jobs to fail. They are no longer failing, and the loop is working again.
- Set up a way to be able to automatically upload both the L, R, Theta plots and the Fitness Score plot to dropbox as we work.
- Set up an email that automatically uploads to dropbox under the folder GP_Antennas/Updates/DailyFitnessScoreImages when you send an attachment with that email.
- We added this command into the loop:
mail -s "Subject" dropbox.2dwp1o@zapiermail.com < FScorePlot2D.png
- It automatically has it upload an image when the loop finishes a generation. It's currently so that the file will be named whatever the subject of the email is so we can control that by whatever is after the -s command. It's currently set to not overwrite.
- Still need to add the Veff plot to dropbox.
- Made gitpush.sh and gitpushforce.sh
- Each time one of us pushes changes to GitHub, it makes a permssion lock on the directories. From now on, after looking at what files are changed (using git status. Remember to make sure to add any data files missed to the .gitignore)users should do:
- git add .
- git commit -m "add message" or git commit --amend --no-edit
- Then run: ./gitpush.sh if you made a full new commit, or use ./gitpushforce.sh if you amended this edit to the previous commit. They both exist in: /fs/project/PAS0654/BiconeEvolutionOSC
- These files run the git push or git push -f commands and then immediately run a chmod to change permissions after.
- Alex is starting the APS slides!
- He should be sharing them shortly. I told him to look at Suren's from last year for guidance.
- We have started working on adding the feature that pulls non-unique (ie repeat parameter) individuals from an antenna database.
- It's currently being developed on the Antenna_Database branch in git.
- Alex has started writing c++ code that reads in the csv file and appends unique items to it.
- Comments from Amy we need to remember:
- Doing "==" on doubles is bad!
- We want this "likeliness" to be between 1 and 10% (start with 10%).
- Remember: is this difference between L stored and new L a fraction of the wavelength?
|
|
|
47
|
Thu May 7 14:35:00 2020 |
Julie Rolla | GENETIS Meeting Update | The students have been meeting to work out the summer schedule. We are dividing into to groups:
(1) Paper writing: Julie, Alex M., Alex. P
- We need Amy to start an overleaf draft (she has the premium version that allows you to share with more than 1 person)
- In tomorrow's collaboration meeting (Now Fridays at 3pm) we are going to request discussion on an outline
(2) Adding wall time variable structure: Eliot, Evelyn, Leo (Alex P. will help oversee)
- In order to prepare group (2) Alex M. has been going over the edits he started and is discussing his code. We also are taking some time to catch them up on the current code -- ie how it runs, what all of the pieces involved do -- so that they can know how to test the code after edits are made, and where each essential piece may lie. We will be going over this for the next few days in good detail.
- Group (2) will then be coming up with a scheduled time each day that they will be coworking on zoom. This will work the same as if they were sitting in an office together. They'll announce this time to everyone once it is set.
Alex P, Alex M, and I are going to push to rapidly finish this antenna database so that the others can create a new branch to work on the wall time variable with. This will leave us very busy for the next few days. |
|
|
48
|
Mon May 11 17:56:36 2020 |
Julie Rolla | GENETIS Meeting Update | We met today for a long time and pushed to catch up. The following is what we have done:
(1) Worked for a bit on our current tasks. They will work on these current tasks before they start on their summer projects.
- Antenna database (Julie, Alex P., Alex M.)
- Worked on a paper outline (Julie, Alex M.)
- Worked on wall time variable (Eliot, Evelyn, Leo)
Note I also have to: (1) Fix merge conflict and make sure students are good about only editing the dev branch moving forward. (2) Get branches and GitHub set up for both the asymmetric bicone and the non-linear bicone.
(2) Went over the details of how the loop works as a group.
- Went over how each piece works.
- Went over how to run it.
- Went over best ways to test new edits.
(3) Went over summer tasks.
- Eliot: Add parameters to bicone (ie make bicone asymmetric)
- Julie & Alex M.: Paper, and helping others
- Alex P.: Frequency database for AraSim (to speed up AraSim)
- Evelyn & Leo: Make non-linear bicone (wiggly sides)
|
|
|
22
|
Sat Nov 30 22:04:30 2019 |
Julie Rolla | GENETIS Loop Work: Fixing Consol Errors | GENETIS Loop Work: Fixing Consol Errors By Julie Rolla
1. Python programs are not running: I think either (1) the user isn't module loading python, or (2) they are loading the wrong version of Python (remember there's a discrepancy for Python2, and Python3).
Solution: I added a line "module load python/3.6-conda5.2" in our araenv.sh script to address the issue. All of the users are sourcing the araenv.sh script in their .bashrc when they login. It's in the directory: /fs/project/PAS0654/BiconeEvolutionOSC. I also added a line in the XF_Loop.sh script (at the top in the "Initialization of Variables" section) that sources araenv.sh just in case the user hasn't put that in their .bashrc. The line reads: source /fs/project/PAS0654/BiconeEvolutionOSC/araenv.sh
Error message fixed:
0
Data successfully read. Data successfully written.
Fitness scores successfully written.
Fitness function concluded.
rm: cannot remove 'runData.csv': No such file or directory
Unable to parse the pattern
^CTraceback (most recent call last):
File "LRPlot.py", line 99, in <module>
plotLR(generations, lengths, radii, g.numGens, g.destination)
File "LRPlot.py", line 45, in plotLR
plt.show()
File "/usr/lib64/python2.7/site-packages/matplotlib/pyplot.py", line 143, in show
_show(*args, **kw)
File "/usr/lib64/python2.7/site-packages/matplotlib/backend_bases.py", line 108, in __call__
self.mainloop()
File "/usr/lib64/python2.7/site-packages/matplotlib/backends/backend_gtk.py", line 83, in mainloop
gtk.main()
KeyboardInterrupt
2. rm runData.csv error: The following error was printed to consol
rm: cannot remove 'runData.csv': No such file or directory
I found these line in section (E) of the loop.
cd "$WorkingDir"
rm runData.csv
python gensData.py 0
cd Antenna_Performance_Metric
python LRPlot.py "$WorkingDir" "$WorkingDir"/Run_Outputs/$RunName 1 $NPOP
cd ..
# Note: gensData.py floats around in the main dir until it is moved to
I think this error is fine. This is how it works: (Step 1) removes runData.csv, if it exists. (Step 2) Runs gensData.py, which creates a new runData.csv. We wanted to delete it before the .py program was run at the start of the run, because gensData.py appends data to runData.csv, if it already exists. (alternatively if runData.csv doesn't exists it creates it-- which is what we want in this case). That's also why we don't delete it later in the "looping part" of XF_Scripts.sh (part G). We want it to append at this point, so we only delete it in the very beginning to make sure it doesn't append to an old run.
3. cp: cannot stat 'Antenna_Performance_Metric/1.uan': No such file or directory (similarly for 2.uan, 3.uan...etc): This error exists just as the fitness score is being calculated -- ie part E. Here is the print:
cp: cannot stat 'Antenna_Performance_Metric/1.uan': No such file or directory
cp: cannot stat 'Antenna_Performance_Metric/2.uan': No such file or directory
Congrats on getting a fitness score!
File "FScorePlot.py", line 1
-*- coding: utf-8 -*-
^
IndentationError: unexpected indent
Note that we have a few errors here. Right now I am directly addressing the error with the "cp" command. It looks like these files are actually being saved as gen_individual#.uan. For example 0_1.uan. The current line in part E says:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/$i.uan "$WorkingDir"/Run_Outputs/$RunName/${i}.uan
done
However, this is not how these files are being saved. I corrected the first iteration of the loop (ie Part E for generation 0) by editing the line to say:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/0_$i.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}.uan
done
And for the looping part of XF_Loop.sh (ie part G) it has been edited to say:
for i in `seq 1 $NPOP`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${gen}_${i}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}.uan
done
4. Made Number of Neutrinos thrown (NNU) in AraSim's setup.txt file a variable: I've added the following line in part D -- as well as the looping section in part G.
These lines replace the number of neutrinos thrown in our setup.txt AraSim file with what ever number you assigned NNT at the top of XF_Loop.sh in the "variables" section. "setup_dummy.txt" is a copy of setup.txt that has NNU=num_nnu instead of a number. NNU is the number of neutrinos thrown (as seen in setup.txt). This "sed" line finds every instance of num_nnu in setup_dummy.txt and replaces it with $NNT (the number you assigned NNT in the variable section at the top of XF_Loop.sh). It then pipes this information into setup.txt (and overwrites the last setup.txt file allowing the number of neutrinos thrown to be as variable changed at the top of this script instead of manually changing it in setup.txt each time. Command works the following way: sed "s/oldword/newwordReplacingOldword/" path/to/filewiththisword.txt > path/to/fileWeAreOverwriting.txt. Both setup.txt and setup_dummy.txt can be found in the directory: /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/
cd "$AraSimExec"
for i in `seq 1 $NPOP`
do
#This next line replaces the number of neutrinos thrown in our setup.txt AraSim file with what ever number you assigned NNT at the top of this program. setup_dummy.txt is a copy of setup.txt that has \
NNU=num_nnu (NNU is the number of neutrinos thrown. This line finds every instance of num_nnu in setup_dummy.txt and replaces it with $NNT (the number you assigned NNT above). It then pipes this infor\
mation into setup.txt (and overwrites the last setup.txt file allowing the number of neutrinos thrown to be as variable changed at the top of this script instead of manually changing it in setup.txt e\
ach time. Command works the following way: sed "s/oldword/newwordReplacingOldword/" path/to/filewiththisword.txt > path/to/fileWeAreOverwriting.txt
sed "s/num_nnu/$NNT/" /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/setup_dummy.txt > /fs/project/PAS0654/BiconeEvolutionOSC/AraSim/setup.txt
#We will want to call a job here to do what this AraSim call is doing so it can run in parallel
cd $WorkingDir
qsub -v num=$i AraSimCall.sh
rm outputs/*.root
done
5. rm cannot remove 'simulation_PEC.xmacro': No such file or directory error appearing in generations after the first (ie not seen in gen 0, but seen in gen 1+).
Statistics initialized.
Tournament parents selected.
Tournament complete.
rm: cannot remove 'simulation_PEC.xmacro': No such file or directory
I found that it tries to remove it twice. See here:
######## Loop (G) ######################################################################################
# 1. Does steps A-F for each generation
###########################################################################################################
for gen in `seq 1 $TotalGens`
do
# used for fixed number of generations
#gen=0; while [ `cat highfive.txt` -eq 0 ]; do (( gen++ ))
# use for runs until convergence
#should the line below be double indented?
read -p "Starting generation ${gen}. Press any key to continue... " -n1 -s
#Part A
cd $XmacrosDir
rm simulation_PEC.xmacro
cd "$WorkingDir"
./roulette_algorithm.exe cont $NPOP
cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv
##chmod -R 777 $WorkingDir
#we can make a function that does this with the flag $gen
#doing this in part A would mean setting $gen=0
#Part B
# First, remove the old .xmacro files
#when do that, we end up making the files only readable; we should just overwrite them
#alternatively, we can just set them as rwe when the script makes them
cd $XmacrosDir
#I'm commenting out the next two lines. They are written to below (around lines 154-170)
#If we keep them this way then the files have restricted permissions
rm output.xmacro
rm simulation_PEC.xmacro
I have commented the first removal of simulation_PEC.xmacro. If there are any issues with it running, we can go ahead and comment it back in; however, I suspect this is not a necessary line to have added in. A run has not been performed since these edits. So, once it has been tested, we can delete the comments out:
#Part A
#I think these next two lines can be deleted. Its repeated again just below...11/30/19 comment by Julie #cd $XmacrosDir #rm simulation_PEC.xmacro
cd "$WorkingDir"
./roulette_algorithm.exe cont $NPOP
cp generationDNA.csv Run_Outputs/$RunName/${gen}_generationDNA.csv
6. Python Error from FScorePlot.py: The error can be seen in the line below.
Congrats on getting a fitness score!
File "FScorePlot.py", line 1
-*- coding: utf-8 -*-
^
IndentationError: unexpected indent
Congrats on getting some nice plots!
I've commented out this strange line at the start of this python program: -*- coding: utf-8 -*-. This fixed the immediate error, but threw another one when I tried to run it outside of the loop with the command
python FScorePlot.py /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie_11_30/ /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Julie_11_30/ 2
The next error it threw can be seen below.
Traceback (most recent call last):
File "FScorePlot.py", line 79, in <module>
fScores.append(fScorei[2:])
AttributeError: 'numpy.ndarray' object has no attribute 'append'
This is Alex P. I edited the append() and changed it to np.append(fScores, fScorei[2:]) on line 79 in FScorePlot.py
7. rm: cannot remove 'outputs/*.root': No such file or directory: This error comes up right after the AraSim job is submitted in part D.
1011007.pitzer-batch.ten.osc.edu
rm: cannot remove 'outputs/*.root': No such file or directory
Waiting for AraSim jobs to finish...
It's definitely coming from the lines:
for i in `seq 1 $NPOP`
do #We will want to call a job here to do what this AraSim call is doing so it can run in parallel
cd $WorkingDir
qsub -v num=$i AraSimCall.sh rm outputs/*.rootdone
We are currently in $WorkingDir at this point-- which is: WorkingDir= /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop. There is not directory called "outputs" in this location. This is why we are getting an error.Can someone explain what .root files we are trying to remove, and why? I cannot proceed unless someone makes this clear to me, as I do not know what files we are looking to remove (and where they exist/when they are created).
Immediately after that, I get the following error:
Waiting for AraSim jobs to finish...
rm: remove write-protected regular file 'AraSimFlags/1.txt'? y
rm: remove write-protected regular file 'AraSimFlags/2.txt'? y
mv: cannot stat '*.root': No such file or directory
It looks like it is coming from the following line:
cd "$WorkingDir"/Antenna_Performance_Metric
mv *.root "$WorkingDir/Run_Outputs/$RunName/RootFilesGen0/"
However-- again-- it looks like it's trying to move .root files from the directory: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric and put them into: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/$RunName/RootFilesGen0/
It can't move these, because they do not exist where it is trying to take them from. Similarly, the question in bold above would help me with this a lot. From what I can see, we are only utilizing an .txt file output from Arasim. Does Arasim output this to .txt, or are we somehow converting from a .root to a .txt?
This error has not yet been corrected!!!!
Cade's comment here:
The .root files live in BiconeEvolutionOSC/AraSim/outputs/ folder. Thus, we were looking at the wrong directory. They were being removed because it is a giant file (GB size) and are created everytime AraSim runs. It just overwrites itself everytime though so it doesnt add up too much. We can have them deleted later if needed. It's our choice on whether we take it out completely, or redirect to delete the .root files in the correct directory.
8. New errors found after running with these corrections: These need to be fixed!
cp: cannot stat ‘Antenna_Performance_Metric/0_1.uan’: No such file or directory
cp: cannot stat ‘Antenna_Performance_Metric/0_2.uan’: No such file or directory
Congrats on getting a fitness score!
Traceback (most recent call last):
File "FScorePlot.py", line 86, in <module>
Gen = np.zeros(fScores.shape[0]*fScores.shape[1])
I found the cp error. It looks like I was incorrect in #3 about this cp command for the .uan files. I moved all of the currently existing .uan files into a new directory in
/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Antenna_Performance_Metric/UAN
With all of them out of the Antenna_Performance_Metric directory, I did a run from scratch to see what it creates. Here's the outcome: So on generation 1, for individual one its going
1_1.uan
1_2.uan
1_3.uan
ie-- > individual_frequencyStep.uan
So once it hits all 60 frequencies for individual 1, it does all 60 for individual 2 by saying:
2_1.uan
2_2.uan
2_3.uan
...etc.This isn't great because it never actually cares about generation number. In section E (so generation 0) we currently have it as:
for i in `seq 1 $NPOP`
do
#Remove if plotting software doesnt need
#cp data/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/0_$i.uan "$WorkingDir"/Run_Outputs/$RunName/0_${i}.uan
done
and in section G of the looping part (gens 1+) we have:
for i in `seq 1 $NPOP`
do
# Gens data used to create a .csv file for the uan file for gain plotting
# cp Antenna_Performance_Metric/$i.uan ${i}uan.csv
cp Antenna_Performance_Metric/${gen}_${i}.uan "$WorkingDir"/Run_Outputs/$RunName/${gen}_${i}.uan
done
This doesn't seem like it's being output from XF the way we thought it is. I think the easiest way to do this is to edit the xmacros to have it output like:
generation_individual_frequency.uan
and thus
cp Antenna_Performance_Metric/${gen}_${i}_${frequency}.uan
If so, we'd have to make a new variable in the bash script for $frequency to go from 1-60, and have it loop moving all of those. I will be looking into this with Cade tomorrow 12/3/19 (since he is the xmacro script expert). Stay tuned!
Phew....thanks for sticking with me on this long update. |
|
|
207
|
Mon Mar 27 13:10:02 2023 |
Alex M | GENETIS Hackathon Summary and Next Steps | This entry will serve to summarize what tasks we've completed and still need to complete after last week's GENETIS hackathon.
Alex
My focus was on the XF scripting for making the PUEO antenna. Before last week, we had a script which made the geometry of a PUEO-like antenna. The xmacro script has been modified in the following ways:
- Now includes power sources connecting the ridges that are opposite each other.
- Generalized to use variables without hardcoding.
- Reads in data from the output files of the PUEO GA.
- Loops over all read in antennas to set up all of the simulations at once.
At this point, the script for setting up the simulations of the PUEO loop is just about ready for use. The only change to be made is to the frequency list. The script for printing out the gain data is also ready, though it only prints out the gain in theta and phi, not the cross polarization.
Here are some outstanding points that need to be hammered out, though they impede the quality and accuracy of a run rather than the actual functioning of the loop.
- The shape of the ridges in the generated antennas from the GA are considerably different from the PUEO antenna.
- This is likely something that can be fixed by adjusting the constraints--the subspace of the parameter space that leads to PUEO-like ridges may be very small as-is.
- You can see an example of an antenna from the GA attached (viewed from slightly below). It looks dramatically different. This seems to be due to constraints on the height and width being too small, but the ridges are also clearly misshapen.
- The curve on the ridges looks like it needs some work.
- For very PUEO-like antennas, the slopes look reasonable, but they do flare in or out slightly. This is more dramatic for ridges that deviate from PUEO's significantly.
- My best guess is that the parametric form of the curves is placing the curves in a plane that is different from the rest of the ridge. This should be fixable by somehow constraining the curve to a given plane.
- The simulation still only outputs polarization data for theta and phi, not the cross polarization.
- It looks like XF naturally gives us the theta and phi polarization. It's not clear how to get the cross polarization gain (which we think means the gain when the signal is at 45 degrees).
- One idea is to redo the simulation but with the antenna rotated 45 degrees in the azimuth. But I'm not certain that that iwll work.
- Amy indicated that the correct way to do this is by simulating the antenna with just one power source at a time.
- Leaving the VPol source would produce the VV and VH data and leaving the HPol would produce the HH and HV data (HV and VH could be flipped!).
- Currently, the antennas have a fixed opening angle. We'd like to add this as a parameter, though at the moment it might be ok to forego that.
- We need to make sure that the power sources aren't too close together (fixed!).
To do for this week:
1. Modify the script to make two simulations for each antenna, each with only one power source.
2. Adjust the constriants on the parameters to fix the curvature.
3. When we begin a run, we will want to just evolve the height and the inner side length (these determine both the inner side length and the outer side length).
Dylan and Jacob
Dylan and Jacob focused on getting PUEOsim/icemc working, which will be the simulation software needed for the PUEO loop. Here's a summary from Dylan:
- Plotting and bash scripts should be mostly good to go.
- Currently have 4/5 of the PUEO prerequisites for pueoBuilder, and the GENETIS-specific code is ready to implement and test when it's built. I’m trying to build the correct root version one more time before today’s meeting, and if it fails again I’m going to ask Will.
- The main thing left is to get the correct version of root installed, which while annoying, shouldn't take too long
To do for this week:
1. Once PUEOsim is working, run it with the usual settings to get the default performance. It will need to be submitted as a job array (ask Alex for help with this if you need; ask Will for advice on how many neutrinos to run).
|
| Attachment 1: Weird_PUEO_Model.png
|  |
|
|
206
|
Tue Mar 21 17:58:21 2023 |
Alex M | GENETIS Hackathon Day 2 | Today was day two of the GENETIS hackathon. See the previous entry for details on day one.
Alex
I continued working on the XF scripting for the PUEO horn antenna. Amy showed me an actual horn antenna from ANITA in Lisa's office so I could get the dimensions and see how the loads (power sources in XF) were offset. Offsetting the power sources led to fixing the asyemmtry problem from yesterday. I tested offsetting the loads by various heights and found that they all give the same gain patterns except for when they overlap, proving that they were interfering with each other when they crossed.
There is still, however, an asymmetry between the x and y axes (but the gain patterns are now symmetric across either of them). This is more reasonable, though I'm not positive that it should be this way. I attached an image of the gain pattern to show this asymmetry. Note that it is still at 83 MHz, which is well outside the PUEO bandwidth, but higher frequency gain patterns show the same thing (but they have more nodes so they're harder to look at). I attached images from the x-axis, y-axis, and above of the gain pattern. One way to check this will be to compare to the data stored in PUEOsim/icemc and see if they demonstrate similar asymmetries.
My next step is to test the constraints we've decided on using the script. I'll try out a few different values of the parameters and then if everything is working I'll use the GA Ryan and Dylan wrote to generate many individuals and have XF make the models for them. We don't necessarily need to see the simulation outputs from those XF runs, we just need to see if we run into error. Beyond that, we need to figure out how to extract the proper gain data from XF to create the same format that PUEOsim and icemc use.
Amy also mentioned that we should add the opening angle of the horn antenna as a gene. This may be a little tricky but should be doable. Once we are set with the current XF script and changing the data formatting to match what PUEOsim and icemc need, I'll work on adding that as a gene.
Dylan and Jacob
We tried to get IceMC running and able to read in gain files - we failed. Something was wrong with my make files and Jacob’s c++ compiler, so neither of us got a working version of IceMC. We then switched gears to focus on pueoSim. I created a branch on the nicemc github to output a veff.txt in the format of IceMC, so we can reuse the already-made code. Jacob and I also have been trying to install pueoSim, but we’ve been running into many errors and issues from the “C++ Gods”. For the next steps, I will finish the veffective branch and create a pull request. Then, I will work on a branch to make it so pueoSim can read in a variable path to the gain files.
Ryan
Tuesday: Created a new branch on the PEUO repository, version0.0, and placed the repository in the same location as the Ara one for familiarity. I also added an alias to this directory onto the genstudents channel if people would like to add the path to their .bashrc's. I then went through the the Evolutionary_Loop directory and cleaned up any residual Ara based files that were not needed to run this loop. This will make navigating and troubleshooting easier as we finish building. This branch was then commited and pushed to the github and submitted a pull request. |
| Attachment 1: ANITA_Gain_X.png
|  |
| Attachment 2: ANITA_Gain_Y.png
|  |
| Attachment 3: ANITA_Gain_Z.png
|  |
| Attachment 4: Top_Down_ANITA.png
|  |
|
|
205
|
Mon Mar 20 17:32:42 2023 |
Alex M | GENETIS Hackathon Day 1 | Today was the first day of the GENETIS Hackathon. We are spending several days this week specifically tackling the PUEO project to get it into full form (or close to it). Here is a progress update after day 1.
Alex
I am working on the xmacro script for the ANITA antenna. Before today, we had a working xmacro script to make the geometry of the ANITA horn antenna. Now I am trying to modify it to make it 1) more true to the actual ANITA antenna (in terms of which parts are electrically connected) and 2) add in the power source and sensors so that we can get simulation results. I've attached the xmacro script here along with a picture of the geometry and some gain patterns. There are four ridges in the antenna. They are all connected by the outer walls of the antenna. However, only the ones directly across from each other are connected by a load (in this case, a power source, since we simulate as a radiator). So we have two power sources, one connected each "ridge bicone".
In making this, I have generated some gain patterns. I want to test a few cases to see if the results coming from XF seem reasonable, though I don't know what I should expect the gain pattern to look like when I've done things correctly. Here are the options I'll compare:
- Just use one "ridge bicone".
- Use two "ridge bicones" with the walls but just one power source.
- Use two "ridge bicones" without the walls and two power sources.
- Use the full form, with two "ridge bicones", walls, and two power sources.
Case (4) is the one I expect to be correct. However, when I simulate that, I get a slightly asymmetric gain pattern compared to what I expect (I expect symmetry across the "ridge bicones"). You can see from the attached image that there is a strange preference in one corner. My guess is that this has to do with which ridges are set to be ground and which are set to 1V (the potential). Feel free to ask me for more details about these figures/scripts on slack and I'll fill them in on here. I'll clean up the script and add in full test cases later this week.
The image of the antenna I attached is a bird's eye view. The green lines represent the power sources I've connected between the ridges. They do appear to overlap, but my understanding is that they are not actually affecting each other (they should just be a representation of the power sources).
The image of a gain pattern (which is 3D and also a bird's eye view) is for the case of the antenna with both power sources and the walls remaining (the antenna is still shown too, to demonstrate the asymmetry of the gain pattern). This is well outside of the frequency range ANITA cares about (83 MHz), and I haven't looked closely at the higher frequencies yet, but I still am surprised by the asymmetry. I'll update tomorrow with more gain patterns to compare.
Dylan and Jacob
Dylan and Jacob are working on the bash scripts for the PUEO loop. They are taking the parts of the bicone loop that are specific to ARA/bicones and changing them to be specific to the PUEO loop.
All the bash scripting outside of implementing the new software we’re developing should now be complete (we just need to debug when we do test runs). The new GA is implemented into the PUEO loop. We tried and failed to build a version of pueoSim on our own. Will has provided us with a precompiled version and instructions for installing it on our users as well as access to the necessary pueoBuilder repo. Jacob and Dylan are scheduled to go through the installation with him on Thursday at 4pm in his office, M2024.
We have also found that peuoSim currently only outputs root files. So, we will need to create a conversion script or modify pueoSim for when we implement it into the loop. Currently, using IceMC looks to be the easiest way to get a test run going. For the next steps, we have a plan for making IceMC read in a variable file path name to avoid recompiles and issues with parallelization (Alex has given us the path to look at how ARA does it). Then, we will work on installing pueoSim and testing its current viability.
Bryan
Upon checking the existing plotting scripts used for previous GENETIS work, it appears that they can be used for PUEO with minimal changes. In order to test the plotting scripts, sample output data from the PUEO loop is needed, or at least test data that mimics the form of the output for the PUEO loop. After more progress is made with installing an MC package and the PUEO loop can be run to produce test outputs, the plotting scripts can be tested and implemented in the bash script. |
| Attachment 1: ANITA_Antenna_Project.xmacro
|
App.getActiveProject().getGeometryAssembly().clear();
var freq = [83.33, 100.00, 116.67, 133.33, 150.00, 166.67, 183.34, 200.00, 216.67, 233.34, 250.00, 266.67, 283.34, 300.00, 316.67, 333.34, 350.00, 366.67, 383.34, 400.01, 416.67, 433.34, 450.01, 466.67, 483.34, 500.01, 516.68, 533.34, 550.01, 566.68, 583.34, 600.01, 616.68, 633.34, 650.01, 666.68, 683.35, 700.01, 716.68, 733.35, 750.01, 766.68, 783.35, 800.01, 816.68, 833.35, 850.02, 866.68, 883.35, 900.02, 916.68, 933.35, 950.02, 966.68, 983.35, 1000.00, 1016.70, 1033.40, 1050.00, 1066.70]
CreatePEC();
var units = " cm";
num_ridges = 4; // either 2 or 4
// Make the walls and a ridge
build_walls(10,1,30);
// Note that to avoid overlap, we need x0 >= y0
x0=0.04001; // previously 0.04
y0=0.04; // previously 0.04
build_ridges(x0, y0, 0.0, 0.1, 0.06, 0.3, 0.26, 0.1, 0.1, 1);
CreateAntennaSource();
CreateGrid();
CreateSensors();
CreateAntennaSimulationData();
QueueSimulation();
// Try making copies of the ridge
var new_ridges = new EllipticalPattern();
Output.println(new_ridges.getInstances());
new_ridges.setInstances(7);
//new_ridges.clone();
//Output.println(new_ridges);
// Makes the outer walls of the horn antenna
// S is the side length of the bottom of the wall
// a is the coefficient for the linear function the walls extrude according to
function build_walls(S,m,H)
{
// Make the edges to define the square
var edge1 = Line( new Cartesian3D(-S + units,-S + units, 0), new Cartesian3D(-S + units, S + units, 0));
var edge2 = Line( new Cartesian3D(-S + units, S + units, 0), new Cartesian3D(S + units, S + units, 0));
var edge3 = Line( new Cartesian3D(S + units,S + units, 0), new Cartesian3D(S + units, -S + units, 0));
var edge4 = Line( new Cartesian3D(S + units,-S + units, 0), new Cartesian3D(-S + units, -S + units, 0));
// Declare sketches to be made from the edges
var wallSegment = new Sketch();
var bottomSegment = new Sketch();
wallSegment.addEdge(edge1);
wallSegment.addEdge(edge2);
wallSegment.addEdge(edge3);
wallSegment.addEdge(edge4);
bottomSegment.addEdge(edge1);
bottomSegment.addEdge(edge2);
bottomSegment.addEdge(edge3);
bottomSegment.addEdge(edge4);
// Let's start by making the bottom
var bottomCover = new Cover(bottomSegment);
var bottomRecipe = new Recipe();
bottomRecipe.append( bottomCover );
var bottomModel = new Model();
bottomModel.setRecipe( bottomRecipe );
// Add the surface
//var bottom = App.getActiveProject().getGeometryAssembly().append(bottomModel);
//bottom.name = "Bottom square";
// Now we need to extrude the edges to get height
var walls = new Extrude( wallSegment, H + units); // Makes an Extrude
var wallOptions = walls.getOptions(); // Gives the possible options for
// We will use the draft law option to extrude linearly
wallOptions.draftOption = SweepOptions.DraftLaw; // allows for draftlaw
wallOptions.draftLaw = "("+m+"*x)"; // Set the expression for the extrude
wallOptions.draftOption = 4; // 4 indicates we use draftlaw
//Walter - Change the gap type to Extended to get the desired shape
wallOptions.gapType = SweepOptions.Extended; // I actually don't like this when we have x^2, but it doesn't do much for just x
//Walter - Create a shell instead of a solid part
wallOptions.createSolid = false; // This way the shape isn't filled in
walls.setOptions ( wallOptions ); // Sets the settings we assigned above
// Make a recipe for a model
var wallRecipe = new Recipe();
wallRecipe.append(walls);
var wallModel = new Model();
wallModel.setRecipe(wallRecipe);
wallModel.name = "Outer Walls";
wallModel.getCoordinateSystem().translate(new Cartesian3D(0,0,0)); // Makes the model start at the origin
// Set the material for these parts
var wallProject = App.getActiveProject().getGeometryAssembly().append(wallModel); // Adds the model to the project
var pecMaterial = App.getActiveProject().getMaterialList().getMaterial( "PEC" ); // Makes the material available
App.getActiveProject().setMaterial( wallProject, pecMaterial ); // Sets the material
//App.getActiveProject().setMaterial( bottom, pecMaterial ); // Sets the material
}
// A function to build the inner ridges
// Here are the arguments, which are primarily used when making the LawEdges
// Bottom x distance from center
// Bottom y distance from center
// Top x distance from center
// Top y distance from center
// Full x
// Full y
// Full height
function build_ridges(x_0, y_0, z_0, x_f, y_f, z_f, tau, beta, S, m)
{
// build_ridges(0.04, 0.04, 0.0, 0.1, 0.06, 0.3, 0.26, 0.1, 0.1, 1);
// Logarithmic slopes
//var Log1 = new LawEdge( "0.04 + u", "0.04 + 0.02/0.26*u", "1/10*ln((exp(3)-1.0)/0.26*u+1.0)", 0, 0.26); // alpha = z_max/beta
//var Log2 = new LawEdge( "0.04 + u", "-0.04 - 0.02/0.26*u", "1/10*ln((exp(3)-1.0)/0.26*u+1.0)", 0, 0.26);
var Log1 = new LawEdge( ""+x_0+" + ("+z_f+"-"+x_0+")/"+tau+"*u", ""+y_0+" + ("+y_f+"-"+y_0+")/"+tau+"*u", ""+beta+"*ln((exp("+z_f+"/"+beta+")-1.0)/"+tau+"*u+1.0)", 0, tau); // alpha = z_max/beta
var Log2 = new LawEdge( ""+x_0+" + ("+z_f+"-"+x_0+")/"+tau+"*u", "-"+y_0+" - ("+y_f+"-"+y_0+")/"+tau+"*u", ""+beta+"*ln((exp("+z_f+"/"+beta+")-1.0)/"+tau+"*u+1.0)", 0, tau);
// Inner straight slopes
var IS1 = new LawEdge( ""+x_0+" + ("+z_f+"-"+x_0+")/"+tau+"*u", ""+y_0+" + ("+y_f+"-"+y_0+")/"+tau+"*u", "("+z_f+"-"+z_0+")/"+tau+"*u", 0, tau); // a_0 + at, b_0 + bt, c_0 + ct
var IS2 = new LawEdge( ""+x_0+" + ("+z_f+"-"+x_0+")/"+tau+"*u", "-"+y_0+" - ("+y_f+"-"+y_0+")/"+tau+"*u", "("+z_f+"-"+z_0+")/"+tau+"*u", 0, tau);
// Bottom line
var BL1 = new LawEdge(""+x_0+" + ("+x_f+"-"+x_0+")/"+tau+"*u", ""+y_0+"", ""+z_0+"", 0, tau);
var BL2 = new LawEdge(""+x_0+" + ("+x_f+"-"+x_0+")/"+tau+"*u", "-"+y_0+"", ""+z_0+"", 0, tau);
// Top line
var TL1 = new LawEdge(""+z_f+" + "+x_f+"/"+tau+"*u", ""+y_f+"", ""+z_f+"", 0, tau);
var TL2 = new LawEdge(""+z_f+" + "+x_f+"/"+tau+"*u", "-"+y_f+"", ""+z_f+"", 0, tau);
// Outer Straight slopes
var OS1 = new LawEdge( ""+x_f+" + "+z_f+"/"+tau+"*u", ""+y_0+" + ("+y_f+" - "+y_0+")/"+tau+"*u", ""+z_f+"/"+tau+"*u", 0, tau);
var OS2 = new LawEdge( ""+x_f+" + "+z_f+"/"+tau+"*u", "-"+y_0+" - ("+y_f+" - "+y_0+")/"+tau+"*u", ""+z_f+"/"+tau+"*u", 0, tau);
// Inner top line
var ITL = new LawEdge( ""+z_f+"", "-"+y_f+" + 2*"+y_f+"/"+tau+"*u", ""+z_f+"", 0, tau);
// Outer top line
var OTL = new LawEdge( ""+x_f+" + "+z_f+"", "-"+y_f+" + 2*"+y_f+"/"+tau+"*u", ""+z_f+"", 0, tau);
// Inner bottom line
var IBL = new LawEdge( ""+x_0+"", "-"+y_0+" + 2*"+y_0+"/"+tau+"*u", ""+z_0+"", 0, tau);
// Outer bottom line
var OBL = new LawEdge( ""+x_f+"", "-"+y_0+" + 2*"+y_0+"/"+tau+"*u", ""+z_0+"", 0, tau);
// Make the sketches
var straightEdge1 = new Sketch(); // All straight edges (IS1, BL1, TL1, OS1)
var straightEdge2 = new Sketch(); // All straight edges (IS2, BL2, TL2, OS2)
var curvedLog1 = new Sketch(); // Logarithmic edge (IS1 and Log1)
var curvedLog2 = new Sketch(); // Logarithmic edge (IS2 and Log2)
var topRectangle = new Sketch(); // Top rectangle
var bottomRectangle = new Sketch(); // Bottom rectangle
// Add the edges to the sketches
straightEdge1.addEdges( [IS1, BL1, TL1, OS1] ); // Inner straight slope
curvedLog1.addEdges( [IS1, Log1] ); // Right logarithm part
straightEdge2.addEdges( [IS2, BL2, TL2, OS2] ); // Inner straight slope
curvedLog2.addEdges( [IS2, Log2] ); // Left logarithm part
topRectangle.addEdges( [ITL, OTL, TL1, TL2] ); // Top rectangle
bottomRectangle.addEdges( [IBL, OBL, BL1, BL2] ); // Bottom Rectangle
//WALTER - The Elliptical pattern is added as a recipe to the parts
//In this case the location of the center and direction of the normal are simple, but for more complex scenarios, may need to use mroe functionality to find them.
var ePattern = new EllipticalPattern();
ePattern.setCenter(new CoordinateSystemPosition(0,0,0));
ePattern.setNormal(new CoordinateSystemDirection(0,0,1));
ePattern.setInstances(num_ridges);
ePattern.setRotated(true);
var cov = new Array();
cov.push(new Cover(straightEdge1));
cov.push(new Cover(straightEdge2));
cov.push(new Cover(curvedLog1));
cov.push(new Cover(curvedLog2));
cov.push(new Cover(topRectangle));
cov.push(new Cover(bottomRectangle));
var pecMaterial = App.getActiveProject().getMaterialList().getMaterial( "PEC" );
//WALTER - We can loop over all our parts and add them to the project as follows. You can use similar concepts above.
models = new Assembly();
for(var w = 0; w < cov.length; w++)
{
var r = new Recipe();
r.append(cov[w]);
r.append(ePattern);
var m = new Model();
m.setRecipe(r);
m.name = "Test Surface " + (w+1);
//WALTER - Seperate array for the models, though we could just get them from the GemoetryAssembly again
models.append(m);
App.getActiveProject().setMaterial( m, pecMaterial );
}
// Work on the loft
var vertex_position1 = curvedLog1.getPosition(curvedLog1.getVertexIds()[0]);
var vertex_position2 = curvedLog2.getPosition(curvedLog2.getVertexIds()[0]);
var loft = new Loft(models.at(2).pickFace(new Cartesian3D (0, 0, 0), vertex_position1, 0.5), "0.0", models.at(3).pickFace(new Cartesian3D(0,0,0), vertex_position2, 0.5), "0.0");
loft.setPart1(models.at(2));
loft.setPart2(models.at(3));
var r12 = new Recipe();
r12.append( loft );
r12.append(ePattern);
var m12 = new Model();
m12.setRecipe( r12 );
m12.name = "Loft 1";
models.append(m12);
//WALTER - append the assembly to the project, then loop over it to assign the material
var assembly = App.getActiveProject().getGeometryAssembly().append(models);
for(x = 0; x < assembly.size(); x++)
{
Output.println(assembly.at(x));
App.getActiveProject().setMaterial( assembly.at(x), pecMaterial );
}
}
// CreatPEC Function
function CreatePEC() //borrowed from XF demo
{
//Make the material. We will use PEC, or Perfect Electrical Conductor:
var pec = new Material();
pec.name = "PEC";
var pecProperties = new PEC(); // This is the electric properties that defines PEC
var pecMagneticFreespace = new MagneticFreespace(); // We could make a material that acts as PEC and PMC, but in this case we just care about electrical components.
var pecPhysicalMaterial = new PhysicalMaterial();
pecPhysicalMaterial.setElectricProperties( pecProperties );
pecPhysicalMaterial.setMagneticProperties( pecMagneticFreespace );
pec.setDetails( pecPhysicalMaterial );
// PEC is historically a "white" material, so we can easily change its appearance:
var pecBodyAppearance = pec.getAppearance();
var pecFaceAppearance = pecBodyAppearance.getFaceAppearance(); // The "face" appearance is the color/style associated with the surface of geometry objects
pecFaceAppearance.setColor( new Color( 255, 255, 255, 255 ) ); // Set the surface color to white. (255 is the maximum intensity, these are in order R,G,B,A).
// Check for an existing material
if( null != App.getActiveProject().getMaterialList().getMaterial( pec.name ) )
{
App.getActiveProject().getMaterialList().removeMaterial( pec.name );
}
App.getActiveProject().getMaterialList().addMaterial( pec );
}
function CreateAntennaSource()
{
// Here we will create our waveform, create our circuit component definition for the feed, and create
// a CircuitComponent that will attach those to our current geometry.
var waveformList = App.getActiveProject().getWaveformList();
// clear the waveform list
waveformList.clear();
// Create a gaussian derivative input wave
var waveform = new Waveform();
var GDer = new GaussianDerivativeWaveformShape ();
GDer.pulseWidth = 2e-9;
waveform.setWaveformShape( GDer );
waveform.name ="Gaussian Derivative";
var waveformInList = waveformList.addWaveform( waveform );
// Now to create the circuit component definition:
var componentDefinitionList = App.getActiveProject().getCircuitComponentDefinitionList();
// clear the list
componentDefinitionList.clear();
// Create our Feed
var feed1 = new Feed();
var feed2 = new Feed();
feed1.feedType = Feed.Voltage; // Set its type enumeration to be Voltage.
//feed2.feedType = Feed.Voltage; // Set its type enumeration to be Voltage.
// Define a 50-ohm resistance for this feed
var rlc = new RLCSpecification();
rlc.setResistance( "50 ohm" );
rlc.setCapacitance( "0" );
rlc.setInductance( "0" );
feed1.setImpedanceSpecification( rlc );
feed1.setWaveform( waveformInList ); // Make sure to use the reference that was returned by the list, or query the list directly
feed1.name = "50-Ohm Voltage Source";
var feedInList = componentDefinitionList.addCircuitComponentDefinition( feed1 );
feed2.setImpedanceSpecification( rlc );
feed2.setWaveform( waveformInList ); // Make sure to use the reference that was returned by the list, or query the list directly
feed2.name = "50-Ohm Voltage Source 2";
var feedInList = componentDefinitionList.addCircuitComponentDefinition( feed2 );
// Now create a circuit component that will be the feed point for our simulation
var componentList = App.getActiveProject().getCircuitComponentList();
componentList.clear();
var component1 = new CircuitComponent();
var component2 = new CircuitComponent();
component1.name = "Source";
component2.name = "Source";
component1.setAsPort( true );
component2.setAsPort( true );
// Define the endpoints of this feed - these are defined in world position, but you can also attach them to edges, faces, etc.
var coordinate1 = new CoordinateSystemPosition( -x0, 0, 0);
var coordinate2 = new CoordinateSystemPosition( x0, 0, 0);
var coordinate3 = new CoordinateSystemPosition( 0, -x0, 0);
var coordinate4 = new CoordinateSystemPosition( 0, x0, 0);
component1.setCircuitComponentDefinition( feed1 );
... 84 more lines ...
|
| Attachment 2: ANITA_Horn.png
|  |
| Attachment 3: ANITA_Gain_Pattern_Both_Sources.png
|  |
|
|
132
|
Tue Apr 6 18:00:23 2021 |
Julie Rolla | GENETIS Google Drive with Talks/Posters, Grant writings, Papers | https://drive.google.com/drive/folders/1iDamk46R2_oOLHtvsOg4jNy05mCiB7Sn?usp=sharing |
|
|
50
|
Tue May 19 12:20:40 2020 |
Alex Patton | GENETIS Daily Updates | Today's Summer 2020 daily update:
As a note, today OSC was down so productivity was more limited
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Mostly just wrote more on the paper in the Genetic Algorithm section. I added some citation that we used in ICRC but there are still more places that should have citations.
|
I might check tonight when OSC is back up to try to push in more updates to the loop because I wanna get Evelyn and Ryan started on running the loop. Putting in those fixes is a big priority because we want to be able to correct the potential issue with XF simulation folders being overwritten and thus uan data not corresponding to the write individuals. The two big things for me in the loop are getting the simulation data to save correctly (and also putting that in the database) and testing that we can replicate results using the specific seed. I'll probably only focus on loop stuff tomorrow.
|
| Alex P. |
Got up before OSC was down to check progress of overnight run, it seems to have worked but I noticed a problem with the database that it wasn't writing to it probably due to a permissions issue but I would have to run another time to see. Shouldn't have affected data but just the use of the database. Run got up to 8 generations with non-zero fitness score which is positive and seems to have fixed the error we originally encountered. Talked to Eliot about pointers and possible errors but was unable to look at the specific error because it is on OSC.
|
Tomorrow plan is to continue to work on database functionality and continue run to get more generations, also want to add the ability to add more plots than just the fitness score to the dropbox automatically. Plots: upload all plots (Fitness, LRT, vEff), remove legend, upload penalized red/green plot too, take off legend, add units to Fitness |
| Leo |
|
|
| Eliot |
Read about pointers and vectors in C++. Talked to Alex P a bit, and have some ideas of things to change to get the GA running. Began reading about antennas. Mostly a down day due to OSC being down.
|
Will implement changes to GA and continue familiarizing myself with how XF reads these values.
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
Draft
|
Thu May 28 15:11:35 2020 |
Alex Patton | GENETIS Daily Update 5/28/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
I wasn't available Thursday or Friday morning.
|
My goal on Friday is to help Alex P. more with finding the cause of the 0 effective volumes we're getting (and also the high effective volumes for small antennas). |
|
Alex P.
|
Set up AraSim to use event read in, got 100 events generated from AraSim in the format of an EventReadIn file for AraSim so that way we have events that are realistic. Tried running and got a run up to the AraSim phase but had some problems with the proper call and order of argumetns. I looked through the AraOut files and thought I got it set up but got some "cannot open" errors I will deal with later. Then hopped on call with Julie, Eliot, and Leo to deal with a Git conflict |
Follow up on AraSim read in to get it functioning properly and then make sure to find events that pass for good antennas so that we can find errors in our 0 vEff antennas. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
d |
|
|
49
|
Mon May 18 15:13:13 2020 |
Julie Rolla | GENETIS Daily Update | Today's Summer 2020 daily update:
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
(See Alex P. for details on working on fixing loop issues)
Went over running the loop with Ryan. Worked with Ryan on trying to get the X11 forwarding for XF to work, but we couldn't get the license. Steven suggested that we try running from a Pitzer desktop, which I think is a good idea if the problem is with the X11 forwarding; if it's a license issue, then we need to figure out where the license is located. I thought this would have been written somewhere in our code, but I couldn't find it, so I may need to email OSC.
Worked more on the symmetric bicone part of the paper. I think I could afford to add detail to the results portion. I included the plots from the APS presentation, but we still need to finish a rerun with the same number of generations and individuals to see if the data makes sense (particularly, I want to see if there was a mistake made when continuing the run after it had finished the 10th generation because I'm skeptical about the fact that we got such large angles in generation 12).
|
I'll meet with Evelyn and Ryan tomorrow to discuss running the loop in more detail. I'll help Ryan use the Pitzer desktop, which will hopefully work. If all goes well, we can get them started on running the loop to check against the data from the run I mentioned in the paper.
I'm going to try writing in the section for Genetic Algorithms in the paper, since Kai said he wanted me to take the first attempt at it. I think for now I'd like to get comments on what I've written on the symmetric bicone portion before I clean it up, especially on whether or not I've given enough detail (and clearly enough).
|
| Alex P. |
Continued to test the database functionality. In the process of testing and debugging we also tested random seed functionality of the genetic algorithm as we used a fixed seed and successfully got repeat results across multiple runs, which allowed better testing as we could be certain to encounter the same scenario when testing and figuring out how to overcome an error.Altered some of the xmacros and xf calls to work with database and created the new macro skeleton for output.xmacro to use just with the database, in the process of looking through the xmacros we found some segments of the code to investigate further. Currently, we just use values 1 to NPOP of the XF simulation and rewrite the contents of 1 to NPOP each generation, but more simulation values continue to be generated even if unpopulated. Also looked more into this and started implementing a way to take out the excess directories while also planning out a way to have each generation use new values rather than overwriting so that we can have a full record of all the simulations across generations rather than just the most recent generations. |
Going to continue to test database, have a run starting gen 1 now and going to continue that tonight, hopefully will find useful results and if not then things to debug. Eliot and Leo asked for help tomorrow with C++ code as they have had some problems with pointers and I agreed to help them tomorrow. |
| Leo |
I, along with Eliot, worked on implementing the new parameter. We have been working on 2 methods, one being adding a 4th gene and the second being adding another chromosome. We are focusing our attention on the chromosome method because we feel it will lead its way nicely into our future work later this summer with adding more complicated parameters. We are running into some issues with pointers and vectors within the GA that we worked on fixing. |
Tomorrow we hope to fix the issues with the pointers and vectors, and hope to test it with multiple generations. |
| Eliot |
Currently working on developing GA further to properly accept 2 chromosomes. The first chromosome has some radius, length, theta and the second chromosome has the same radius and theta, but its own unique length. I currently have the code properly populating an initial pool. However, it can not quite evolve. The GA is free of syntax errors, but I am getting some type of pointer error. I am working to resolve this. I believe it is the only obstacle left on the GA side of things, but it may not be.
|
I will continue working on fixing the errors in the GA, and hopefully finish that tomorrow. If so, I will begin learning about the XF implementation.
|
| Evelyn |
Had class, so I couldn't meet with the group
|
Get caught up with the stuff from today |
| Ryan |
|
|
Julie: Since I won't be doing GENETIS work every day necessarily, I'll add myself down here with a blurb when I am working with the others. I will also be continuing my daily updates on Dropbox for non-GENETIS specifics. |
|
|
52
|
Wed May 20 10:59:29 2020 |
Alex Patton | GENETIS Daily Update | Today's Summer 2020 daily update:
Learn how to use the final part of the loop
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Currently running the loop without the database to change the fix Alex and I made for XF to properly place data in the simulation folders. It looks to us like this was a problem that needed to be fixed. I'm running with the specific seed set in the roulette algorithm so we can replicate the results. I showed Evelyn and Ryan how to run the code today. Ryan was able to get a Pitzer Desktop, so until we can figure out how to get his X11 forwarding to work for XF, that will be a temporary fix. The only drawback is that Pitzer desktops require a GPU, so wait times can be high. I started this run by having him screenshare on zoom and then guiding him through running the loop. We got through the submission of the first batch of XF jobs (and also found some errors, so we had a chance to demonstrate fixing those).
|
My goal is to get several generations of this run done so that I have enough data to see if my fix worked. If so, I'm going to implement it in the database version and we'll perform a test with that. I'm meeting with Evelyn and Ryan at 1:30 to discuss the loop again, so I'll try to have a run going near where we left off today so they can see the rest. Once we know that we've fixed this XF problem we should be ready to fully implement the database by merging with the master branch.
|
| Alex P. |
Implemented some changes to get graphs working and automatically submitted all 5 graphs requested for to dropbox, then helped Alex M debug his changes and helped him show Evelyn and Ryan the loop. While Alex M ran to test the changes, helped Eliot and Leo with their pointer problem and fixed it so it doesn't give an error but they still need to make some changes to get it functioning how they want. |
Continue to help Alex M with fixing the XF simulationNumber issue and make sure the graphs are all valid and uploading properly. After we get confirmation that our XF fix is working we will implement it to the database version too. |
| Leo |
Me and Eliot continued work on the 2 chromosome method. We were successful at fixing our pointing/vector issue, but we have a new error with the data_read function within the GA. |
Tomorrow we will meet with Julie to flush out the issue with data_read. |
| Eliot |
Continued working on the erros with the roulette alg. I have gotten the code to a running state, however the data is not outputting correctly. I believe this is an issue within the roulette function or dataread function.
|
I will further investigate roulette and dataread function and meet with Julie to do so.
|
| Evelyn |
Learned how to run the updated version of the loop and about the database, but we weren't able to get through an entire generation.
|
Learn how to use the final part of the loop. |
| Ryan |
I found a workaround (using a Pitzer desktop until I can fix the error with my machine's Linux subsystem) to my previous issue and was able to do two runs of the loop. |
Alex M. Plans on helping me finish learning how to run the loops.
|
|
|
|
53
|
Thu May 21 14:31:08 2020 |
Alex Patton | GENETIS Daily Update | Today's Summer 2020 daily update:
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Continued the run testing the XF fixes with Alex P. We think our fixes are working, but we want to get a few more generations through. We checked the uan files and saw different data, which we didn't see on the past runs, so this indicates the fix is working. Alex and I also showed Ryan and Evelyn more of the loop and how to interpret the plots being generated. I also put in the red-green color plots, but we need to get through the next generation to see if they work.
|
I'm going to try to get through more generations tonight so we can see what other fixes we still need to implement. I'll continue looking at any bugs that arise tomorrow, but we're getting very close to being able to mergre branches.
|
| Alex P. |
Picked up Ryan and Alex's run from yesterday to progress to the end of gen 1. Found an error related to the changes and moving UAN files after gen 0 with proper naming. After error was fixed went back to run and made sure the graphs all worked. XFintoARA.py had to be altered to fit the new changes. We saw that the UAN files now differed between individuals across generations whereas they hadn't before. Worked with Alex M and showed Evelyn and Ryan more about the loop and the graphs while also debugging our problems while sharing with them so they can see the process when running the loop and running into an error. |
Continue to run, hoping to iron out all errors so it can run on its own. Along with that making sure all the graph tools work, currently have an error in the Red/Green penalized version and also in the automatic upload of some graphs. Also hope to continue the old database run past generation 9 before implementing these changes on the database version. |
| Leo |
|
|
| Eliot |
Font Continued to work on erros within roulette algorithm. Met with Julie to discuss the issue we were still having. Soon after meeting, Leo and I got it to a near perfect state. There are only a couple genese that were unique that shouldn't have been. This is likely a small error that I will look into tomorrow.
|
I plan to finalize the roulette alg to accept 2 chromosomes with 3 genes a piece. These will have shared radius and theta but unique lengths.
|
| Evelyn |
Learned how to run, debug, and read the graphs in the second half of the loop, so that I can start getting some real data as soon as the loop is ready to be run again. |
|
| Ryan |
|
|
|
|
|
46
|
Sun Apr 19 21:16:21 2020 |
Alex Machtay | GENETIS APS Slides | Here's the presentation we gave for the April APS meeting today. Atttached is the power point of the presentation and a pdf version. |
| Attachment 1: Genetis_APS_Presentation.pptx
|
| Attachment 2: Genetis_APS_Presentation.pdf
|
|
|
176
|
Tue Aug 9 11:36:24 2022 |
Ryan Debolt | GA User guide (pdf) | |
| Attachment 1: VPol_GA_User_guide.pdf
|
|
|
143
|
Fri Feb 4 17:59:41 2022 |
Ryan Debolt | GA Updates | The following plots are ittereations if the test loop that add increasing improvements to the GA.
The first plot shows the GA's behavoir unaltered from our previous runs (80% roulette, 20% tournament elite selection on).
The second plots shows when we use 90% rank selection and 10% tournament, elite selection off.
Plot 3 shows when we add an offset to restrict the values of the fitness function to be more within the range of the main loop.
Plot 4 shows when we add a gaussean mutation function that is applied to crossover individuals (rate and gaussean width chosen by guess).
The following are papers I have looked at while modifing the GA (not nessisaraly recently).
https://pdfs.semanticscholar.org/5733/418cbf21dedc9e5c04351ded4a989f1ff67e.pd
https://www.sciencedirect.com/science/article/abs/pii/0165607493902157
https://www.scientific.net/AMM.340.727
http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.28.1400&rep=rep1&type=pdf
https://arxiv.org/pdf/2010.04340.pdf
https://d1wqtxts1xzle7.cloudfront.net/30694440/10.1.1.34.9722.pdf?1361979690=&response-content-disposition=inline%3B+filename%3DUsing_genetic_algorithms_with_asexual_tr.pdf&Expires=1612908683&Signature=X93Gsc47AS0xqWf1SPLjG~7sNkoXSOXfnq1GpZ2QaPrYw9x9mWwASStW2IWexo7QBzbkhzcE5tZ~CmQA1MHN-paiNFIx2ed8VNS3IhesMnotKM0mSgUZ37BCleHT9BgGkUUum8mTJBAzCUaECn6RYjm1CZpfwVPC9zwuA~DnXBST4pGlQdna22D--sHwXgX~3U3gDUSxqk8mLI0gtn~Xued3XqsTGuMUKwJ2D9UpD5yp42-3IrH6d5CZREjEfXY2geTopQ-uNkr3eOriDj0UZqSrDw5mczmod3kQrQncgd~G2Kyda4RlIs8VDzQs~BGgszHJhSDAuKDrXr8P--9tVg__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA
http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.438.7389&rep=rep1&type=pdf
https://arxiv.org/pdf/2102.01211.pdf |
| Attachment 1: Original_Params.PNG
| 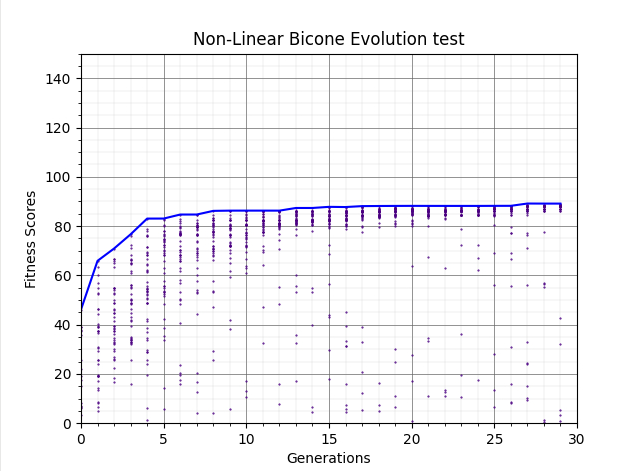 |
| Attachment 2: Rank_params.PNG
| 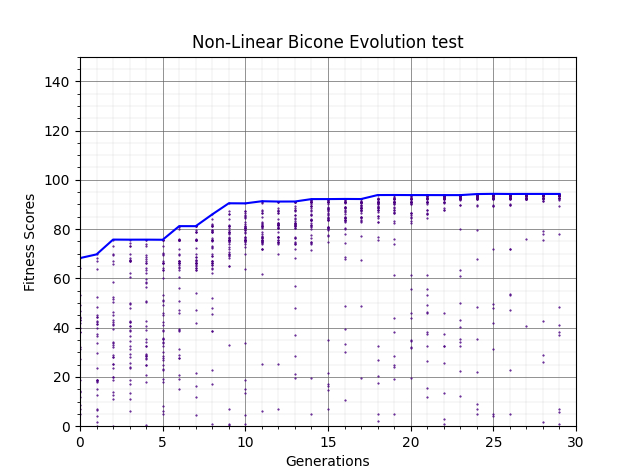 |
| Attachment 3: Range_restriction.PNG
| 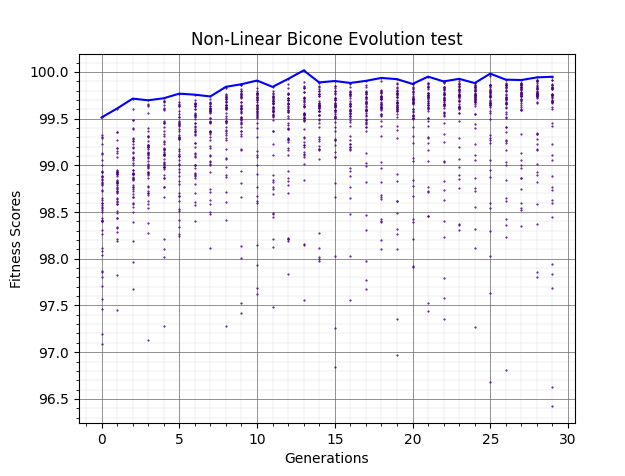 |
| Attachment 4: Mutation.PNG
| 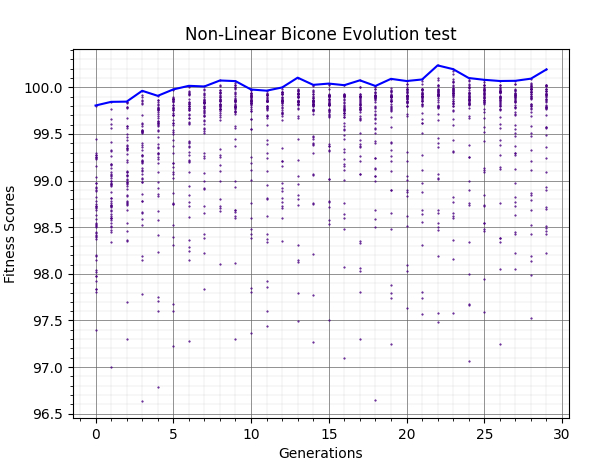 |
|
|
154
|
Fri May 20 14:26:39 2022 |
Alex M | GA Papers | I'm making this entry so that I can record some interesting papers we find on genetic algorithms. Feel free to update this list with links to papers and maybe make a description of what was interesting/of note in the paper.
|
|
|
Draft
|
Fri Aug 21 15:19:10 2020 |
Ryan Debolt | Friday Updates |
| Name |
Update |
Plans for next week |
| Alex M |
|
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
This week I completed work on a set of graphs that shows many instances of of a run type on a graph to show the spread that runs give. I am still working on finishing up the plots that show averages of roulette to tournament for each sigma but the should be finished soon. |
Finish the aforementioned plot. |
| Ben |
|
|
| Ethan |
Ben and I spent the week working on two seperate python scripts, one to plot the fitness scores of each child in each generation, and another to replace dummy Veff scores with Veff scores found in another file. We also updated the documentation on OverLeaf. |
On monday we will continue to keep updating the documentation as well as continueing to imporve AREA. |
|
|
|
125
|
Fri Dec 11 17:47:48 2020 |
Ethan Fahimi | Friday Updates |
| Ethan F |
Fixed the issued AREA was having with finding test_{ind}.txt, now to fix problems with finding Veff and the project should be working. |
| |
|
| |
|
| |
|
| |
|
| |
|
| |
|
|
|
|
155
|
Fri May 20 17:17:32 2022 |
Ryan Debolt | Fitness Functions Test | Bellow lies plots testing different scores and comparing them using a chi^2 score.
The functions used are as follows
Gaussian: e^(-2) (Red)
Inverse: 1/(1+(O-E)^2) (Purple)
Algebraic: 1/(1+(chi^2) )^(3/2)) (Green)
Chi: 1/(1+chi^2) (Blue)
Which are plotted here:

|
| Attachment 1: Alg_Chi.png
|  |
| Attachment 2: Alg_fitness.png
| 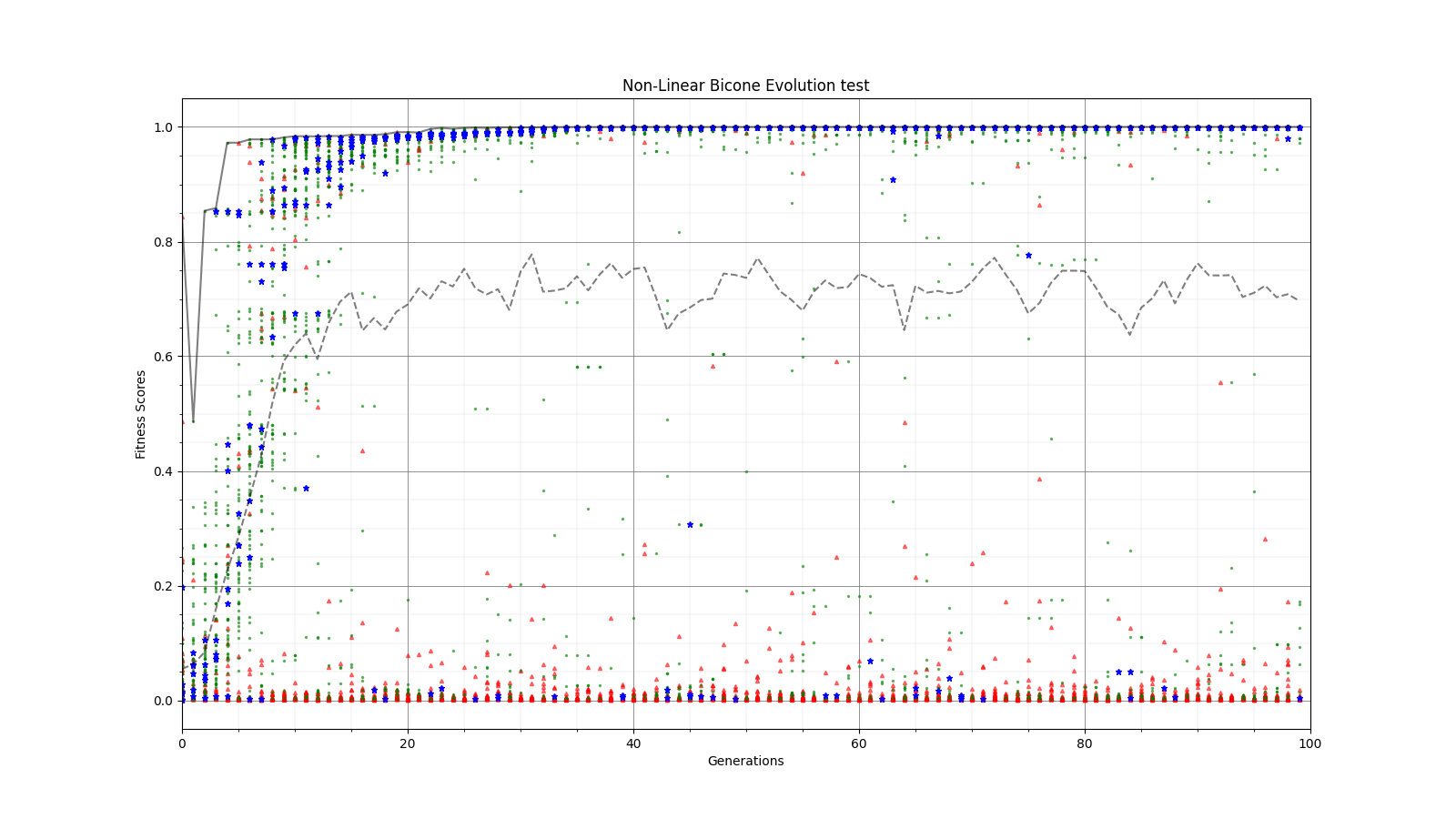 |
| Attachment 3: inv_Chi_Fitness.png
| 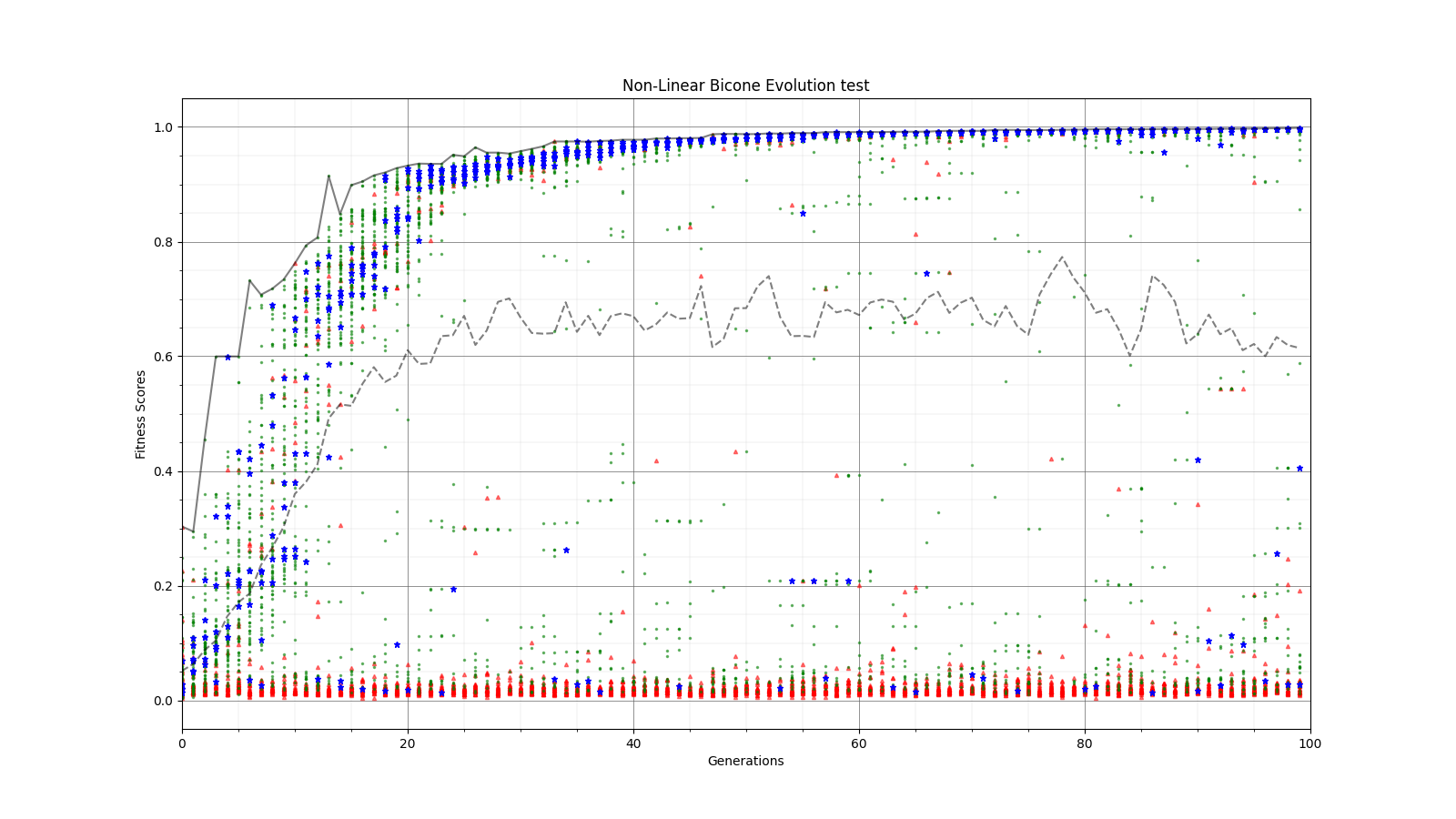 |
| Attachment 4: Inv_Chi_Chi.png
| 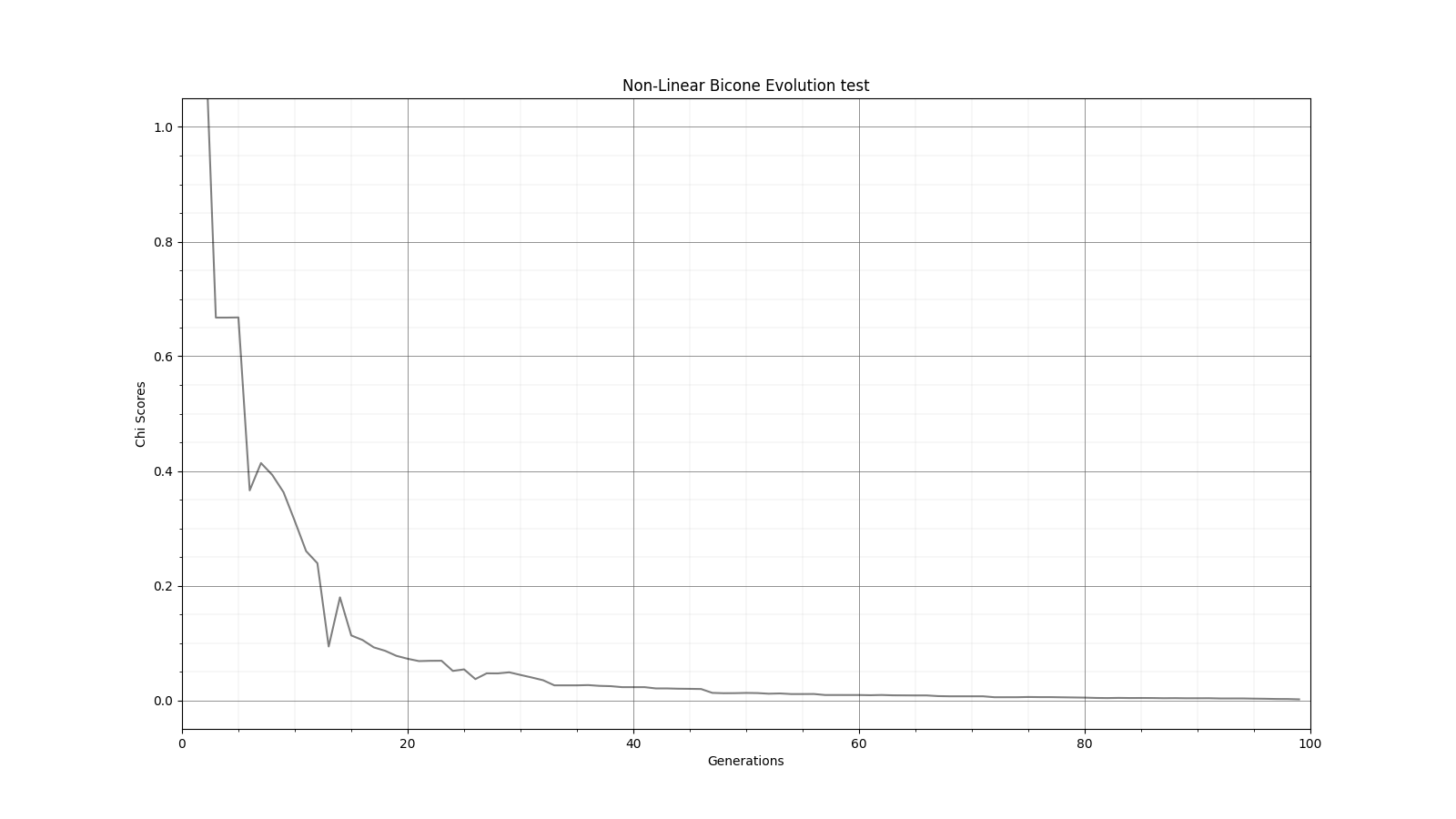 |
| Attachment 5: Gauss_Chi.png
| 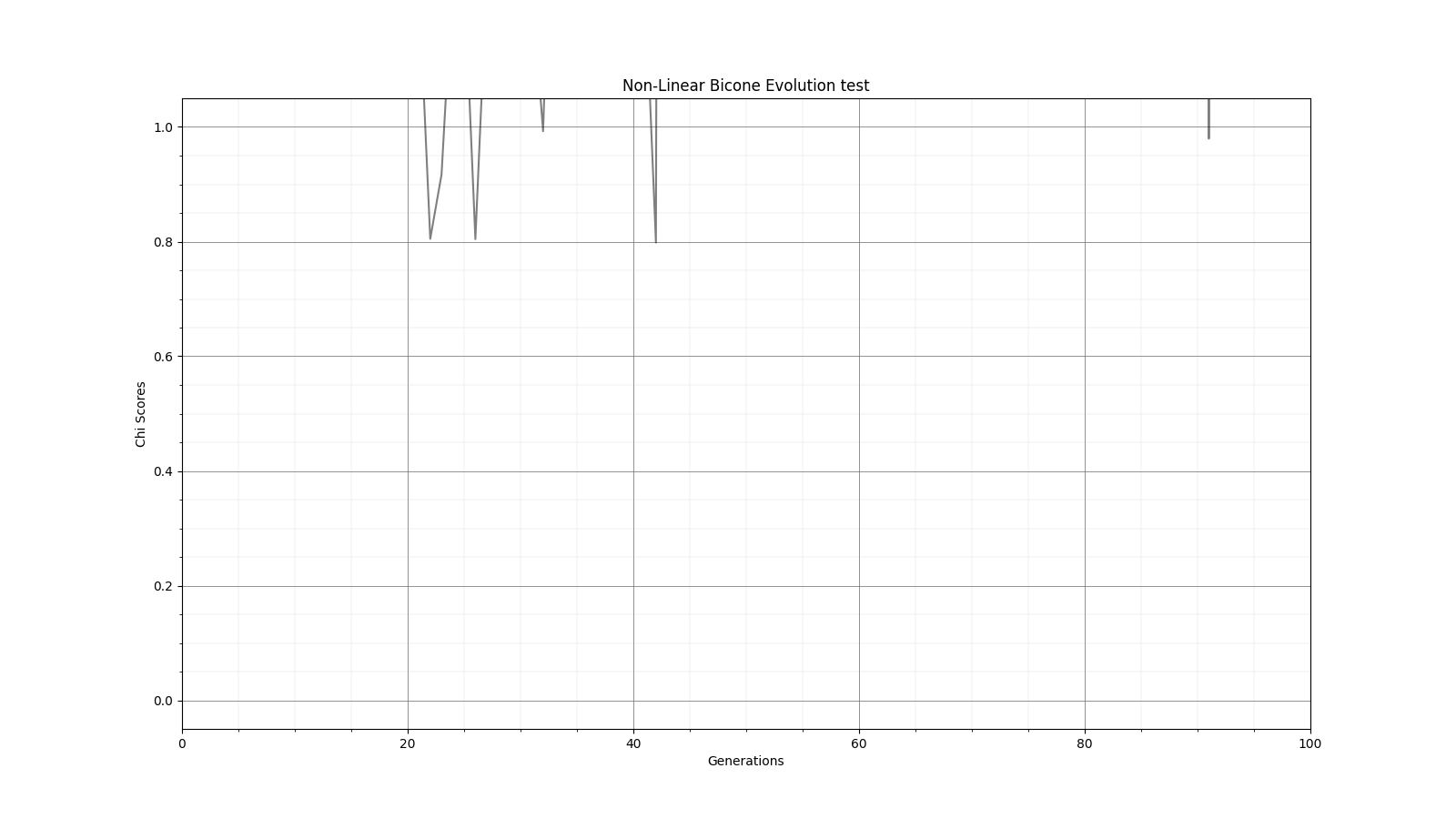 |
| Attachment 6: Gauss_Fitness.png
| 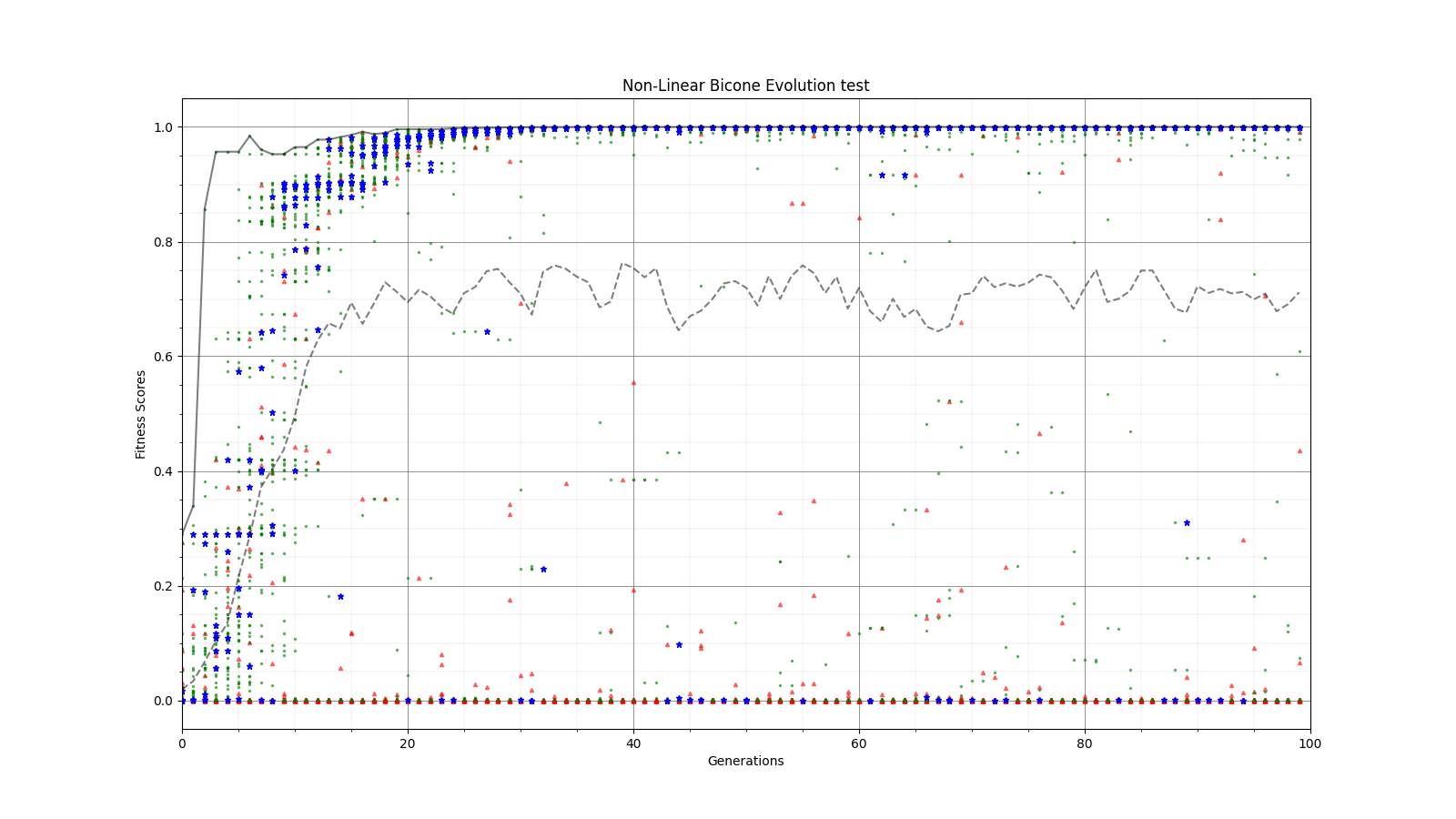 |
| Attachment 7: Inv_Chi.png
| 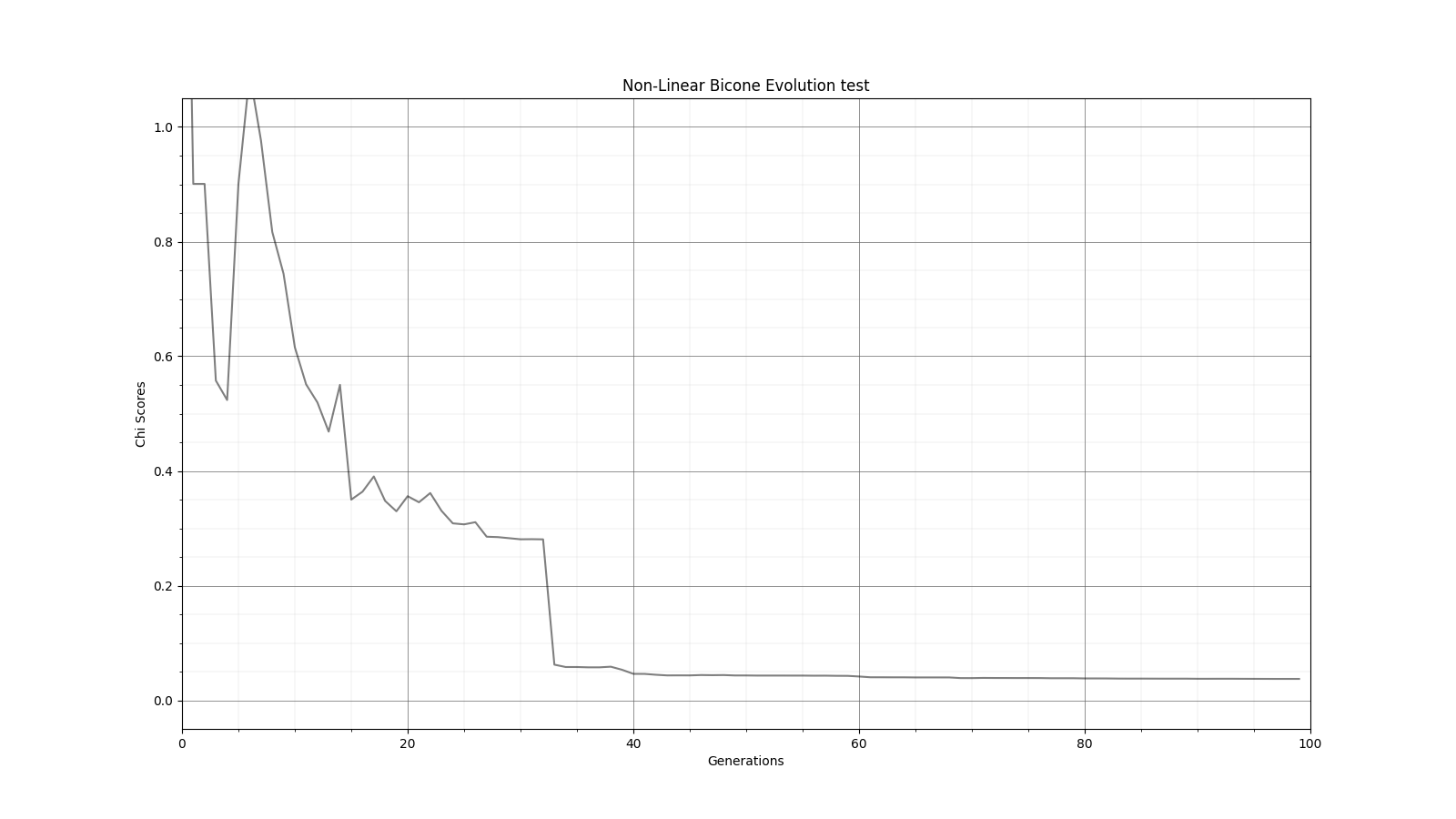 |
| Attachment 8: Inv_Fitness.png
| 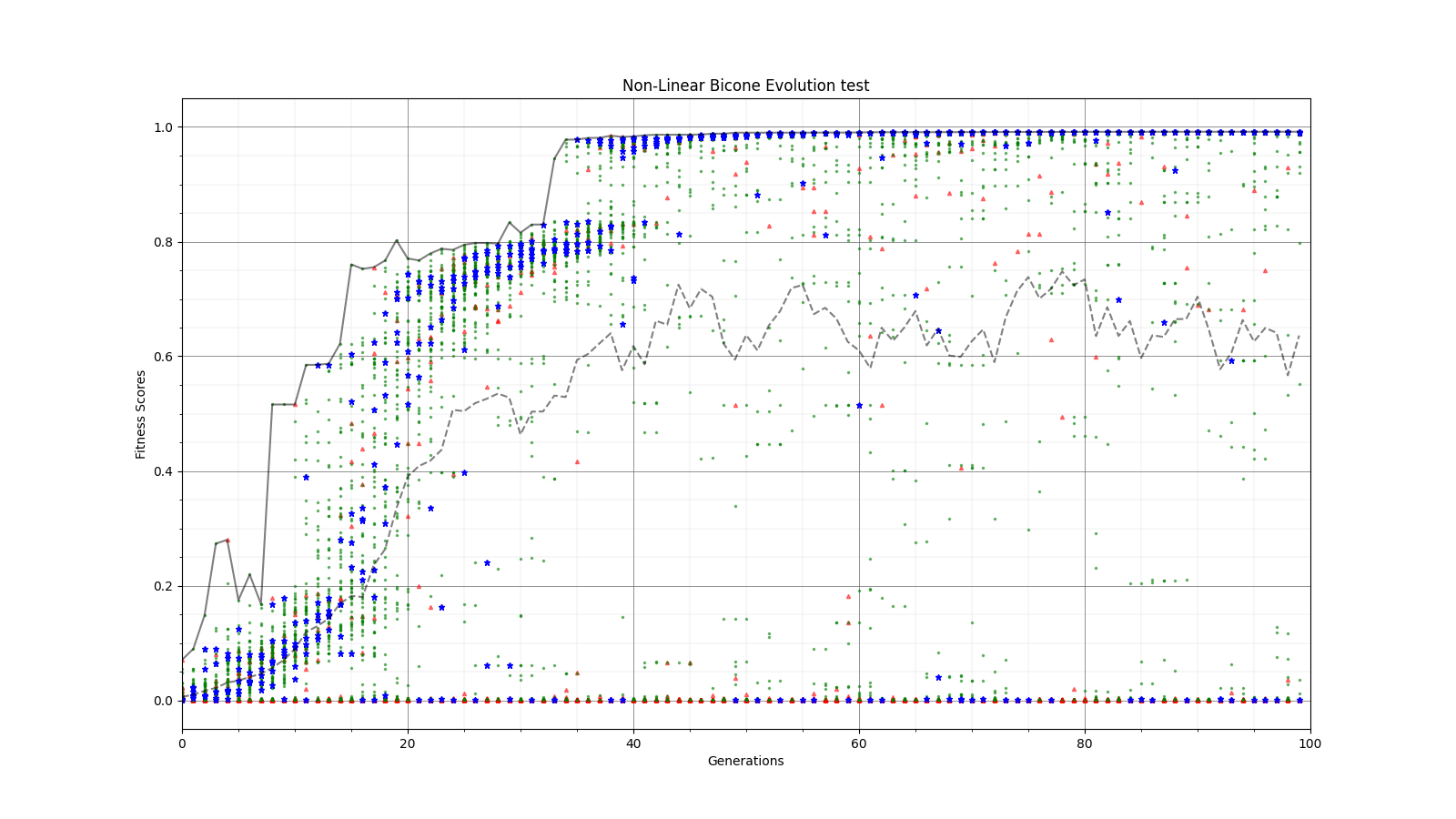 |
|
|
105
|
Fri Aug 28 16:31:56 2020 |
Alex M | First Results Slides | Here's a power point I showed today at the GENETIS meeting for the two most recent runs we did. The first run was symmetric and we ended it at 15 generations. The second one is ongoing and is shown up through generation 9 on the power point. |
| Attachment 1: GENETIS_Meeting_Loop_Results_2020_08_24.odp
|
|
|
209
|
Tue Apr 4 13:51:56 2023 |
Alex M | Final To Do List for PUEO Loop | At the main GENETIS meeting yesterday, we collected the last few things that need to be worked out in order to get the PUEO loop in working order. Here's a list
-
Skeletonize the xmacros (Alex) 04/10/23 In progress
-
We need to modify PUEOsim to read in different gain files (Dylan) Changes made 04/10/23 Testing to be done.
-
Currently, PUEOsim reads in files like vv_0, vh_el, etc. It needs to be modified to take in the gainfiles as arguments and read different ones for each individual.
-
Requiers slight modification of the batch job scripts, but these are relatively small
-
Slight modification on the fitness score read in script (Dylan) Changes made 04/10/23 Testing to be done.
-
Fix Ezio's comments on XFintoPUEO.py and modify to read in outputs from two files (Jacob) 04/10/23 in progress, further work this week
-
Since we are running XF twice (once for each power source), we need the conversion script to read in from two files and print out to 8 files (vv_0, vh_el, etc)
|
|
|
16
|
Tue Nov 5 16:19:20 2019 |
Julie Rolla | Fall update | We are off again with a new team. The new team consists of Alex Machtay, Alex Patton, Cade Sbrocco, Eliot Ferstl, Evelyn Shank, Mitchell Halley, Scott Janse. A spreadsheet of tasks and deadlines can be found here: https://docs.google.com/spreadsheets/d/1CkcdbediFbripdlwPw6UmhAHdAl5IdkL2efaseJkTTM/edit#gid=0
Updates:
We are now running on OSC. We have acquired a version of XF on OSC and have now transferred the loop to OSC. All of the xmacros for XF should be functioning, and we have managed to make the bash script auto import the xmacro scripts to XF when it boots. The loop exists on /fs/projects/PAS0654/BiconeEvolutionOCS. This is a shared copy so that everyone can run off of the same installation. This is important so that we can make sure everyone is pushing edits regularly, and so that everyone is always running with the most up-to-date edits.
The loop had not yet been tested for it's current status. Alex M. is testing it and we are waiting to hear back.
Tasks:
- Check continuity of loop
- Update: Alex M is testing this
- Install Arasim to /fs/projects/PAS0654/BiconeEvolutionOCS to be edited and messed with
- Update: this has been completed as of 11/5/19
- Test Alex M's code for editing Arasim's Setup.txt variables
- Clean bash script
- Update: Mitchell is working on this.
- Make version of bash script which writes .job scripts for Arasim
- This must write a different .job for each individual so it parallelizes Arasim running for each individual
- The number of .jobs written must depend on $NPOP
- ie this bash script should be writing .jobs from scratch each time as (1) the number of them we need depends on $NPOP, (2) the names of the input and output files depends on $NPOP.
- It should also delete .jobs at the end -- ie when all generations have completed so that we don't have a random number of .jobs floating around. For example, say we run with 10 individuals one time, and it writes 10 .jobs ($NPOP=10), and then the next run we only have $NPOP=5, we will have 5 extra .jobs floating around not being used unless we clean them all at the end line of the run in the bash script.
- Update: Work on this has begun by Alex M, Cade, and Evelyn (Alex P has also been messing with Arasim).
- Make XF be submitted as a job
Students have been posting individual updates here: https://www.dropbox.com/home/GP_Antennas/Updates
|
|
|
62
|
Wed Jun 3 15:20:04 2020 |
Julie Rolla | Existing Info for Current Projects | Paperclip Antenna (Ryan and Evelyn working on it):
Asymmetric Bicone (Leo and Eliot working on it):
AREA (Ben working on it):
- Github? We need to ask Steph where their past work exists.
- See AREA thesis attached
Symmetric Bicone(Alex M. and Alex P. working on it):
|
| Attachment 1: AREAthesis.pdf
|
|
|
223
|
Fri Jun 2 00:21:36 2023 |
Ryan Debolt | Error test results | Attached is a plot containing bar graphs with error bars representing the average number of generations it took for the GA to achieve a chi-squared value of 0.25 (roughly equated to a 0.8 out of a max 1.0 fitness score). Unlike the fitness scores used by the GA, these values do not have simulated error attached to them and are therefore a better measure of how well the GA is optimizing. These results were obtained by running 10 tests in the test loop for each design, population, and error combination and solving for the average number of generations to meet our threshold and the standard deviation of those scores. From this, a few things immediately pop out, I will address the more obvious one later. But essential for this test, we can see that increasing our population size seems to have a more direct impact on the number of generations needed to reach our threshold than decreased error does in both designs. My best guess regarding this is that the GA depends on diversity in its population in order to produce efficient growth, and an increase in the number of individuals contributes to this, allowing the GA to explore more options.
This leads me to the more easily spotted trend; PUEO is much slower than ARA. This presents an anomaly as this is the opposite of what we would expect from this test loop as PUEO has fewer genes (7) to optimize than ARA (8). It is also important to note that no genes are being held constant in this test for either design, so both designs have the full range of designs provided they are within the constraints. With that in mind, my guess as to what causes this phenomenon is that PUEO's constraints are much stronger than ARA's. How this may affect the growth is that it more heavily bounds the possible solutions, which makes it harder for the GA to iterate on designs. It is possible that during a function like a crossover, the only possible combinations for a pair of children are identical to their parents, effectively performing reproduction. This could limit the genetic diversity in a population and therefore cause an increase of generations needed to reach an answer. We could in theory test this by relaxing the constraints done by PUEO and then running the test again to see how it compares.
Finally, You will notice that no AREA designs are shown. This is because, under current conditions, they never reached the threshold within 50 generations. However, Bryan and I think we know what is happening there. AREA has 28 genes, about four times the amount of genes that our other designs use. Given that our current test loop fitness measure is dependent on a chi-squared value given by: SUM | ((observed(gene) - expected(gene))^2) / expected(gene) |, we can see that given more genes, the harder it gets to approach zero. For example, we can imagine in an ARA design if each index of the sum equals 0.1, you would get a total value of 0.8, while AREA would get a value of 2.8, which seems considerably worse, despite each index being the same. Upon further thinking about it, Bryan and I do not think that a chi-squared is the best measure of fit we could be using in this context. Another thing we thought about is that we have negative expected values in some cases. We have skirted around this by using absolute values, but upon reflection believe this to be an indicator of a poor choice in metric. Chi-Squared calculations seem to be a better fit for positive, independent, and normally distributed values, rather than our discrete values provided by our GA. With this in mind, we propose changing to a Normalized Euclidean Distance metric to calculate our fitness scores moving forward. This is given by the calculation: d = sqrt((1/max_genes) * SUM (observed(gene) - expected(gene))^2). This accomplishes a few things. First, it keeps the same 0 -> infinity bounds that our current measure has, allowing our 1/(1+d) fitness score to be bounded between 0 and 1. Second, it forces all indices to be positive so we don't need to worry about negative values in the calculation. Third, this function is weighted by the number of genes present for any given design, making them easier to compare than our current measure. Finally, as our GA is technically performing a search in an N-dimensional space for a location that provides a maximized fitness score, it makes sense to provide it the distance a solution is from that location as a measure of fit in our test loop. We created a branch on the test loop repository to test this and the results are promising as results from the three designs are much more comparable for the most part (though we still see some slowdown we think is contributed to constraints as mentioned above). Though some further input would be appreciated before we begin doing tests like the one we have done in the plot below. |
| Attachment 1: test_results(1).png
| .png) |
|
|
169
|
Tue Jun 28 12:49:53 2022 |
Dennis Calderon | Effective Volumes from AraSim: Curved Sides and Straight Sides (Paper Run) | Summary of results for 3million event simulations in AraSim with both GENETIS version and more recent {~11/2021) version.
Using errors for effective volume from the .out files.
Example shown below.
Radius: 3000 [m]
IceVolume : 8.4823e+10
test Veff(ice) : 6.50867e+09 m3sr, 6.50867 km3sr
test Veff(water eq.) : 5.96845e+09 m3sr, 5.96845 km3sr
And Veff(water eq.) error plus : 0.543588 and error minus : 0.543588
|
| Attachment 1: AraSim_Summary_Effective_Volumes.pdf
|
|
|
226
|
Sun Jun 18 21:32:03 2023 |
Dylan Wells | Default Toyon Antenna Simulation | To act as our comparison to the evolved antennas while plotting, we have done a simulation of pueoSim with 4,000,000 neutrinos for the measured toyon gains found in /fs/ess/PAS1960/buildingPueoSim/pueoBuilder/components/pueoSim/data/antennas/measured
In order to run the jobs, I used the runJobs.sh script found in /fs/ess/PAS1960/buildingPueoSim which submits job runs of 40,000 neutrinos at a time, mirroring what we do in the actual loop.
The resulting data is now located in /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Toyon_Outputs
Veff: 11750.0503856
Plus Error: 89.0051458227
Minus Error:88.9007824246
The fitness score plots have been updated to read this value in as a comparison.
|
| Attachment 1: 248599981-8eacbc83-d42e-4da4-9746-bda05b2b4a38.png
| 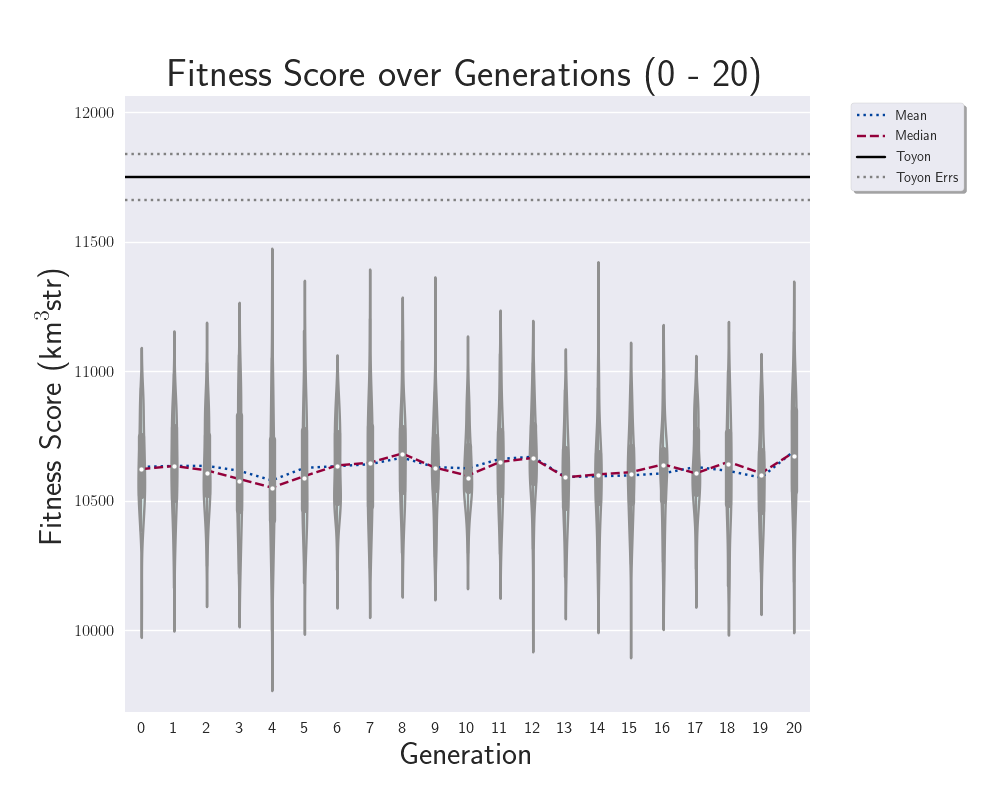 |
| Attachment 2: 248599974-bee7a4db-34c6-4cfd-bde0-3682ff3ebaf7.png
| 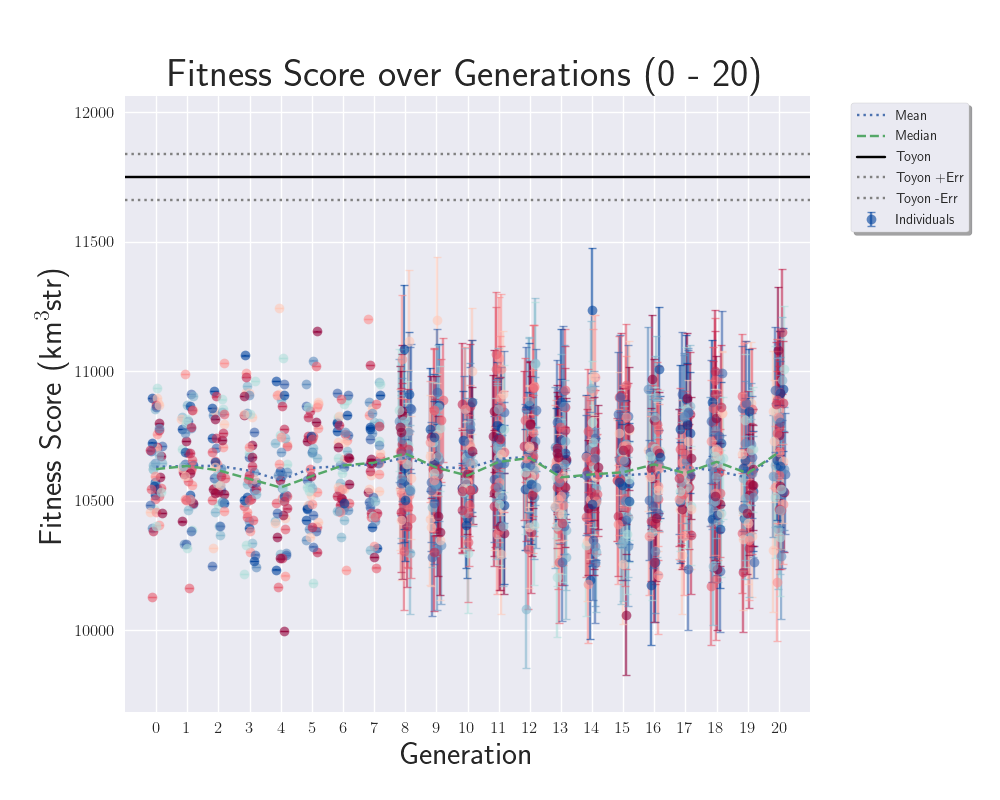 |
|
|
111
|
Mon Oct 5 21:06:29 2020 |
Everyone | Data Runs | Machtay_20200831_Asym_Length_and_Angle 10 individuals
Machtay_20200911_Symmetric 10 individuals, fewer neutrinos
Machtay_20200914_Asymmetric_50_Individuals 50 individuals, fewer neutrinos
Machtay_20200929_Asymmetric_test_2 50 individuals, fewer neutrinos, broaden parameter range
|
|
|
101
|
Fri Aug 7 14:16:01 2020 |
Alex M | Daily Update 8/7/20 |
| Name |
Update |
Plans for Monday |
|
Alex M
|
|
|
|
Alex P
|
|
|
|
Eliot
|
|
|
|
Leo
|
|
|
|
Evelyn
|
|
|
|
Ryan
|
|
|
|
Ben
|
|
|
|
Ethan
|
|
|
|
|
|
100
|
Wed Aug 5 16:08:40 2020 |
Alex M | Daily Update 8/5/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Continued running the loop up to gen 15. I also adjusted the LRTPlot script so that it can slightly spread the points along the generation axis so that you can see overlapping points, but I still need to implement this in the loop.
Helped Ryan design the bash script for running through paperclip many times. It's working and he ran it for a while to get ~1000 outputs checking the performance of different mutation sizes. Next he'll parse through them to find what sigma appears best.
|
Amy suggested that I make it possible for the roulette algorithm to record the mutations and parents for each individual so that we can get a better sense of the history of the evolution, so I'll work on that tomorrow. If it looks to be working, I'll start a run with an asymmetric length. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Completed bash script for automating runs and generated 1100 txt and csv (each) files of runs to be able to see how the ratio of roulette to tournament and standard deviation in the mutation function affect the fitness scores after 100 generations. |
Work on writing a program to go through the txt files and collect the information of the highest fitness score, stored in line 6 of each one, and figure out which combination of parameters gives the highest average. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
99
|
Tue Aug 4 17:30:09 2020 |
Alex M | Daily Update 8/4/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I've continued the run I started yesterday. I ran into a bug where the LRT plots weren't being made but it should be fixed now. The loop is on generation 10, but it hasn't really converged or shown a clear pattern. We'll keep this run going until at least gen 15 probably. |
If we finish this run, I can start another run using the asymmetric version. The goal would be to do a similar number of generations using the asymmetric length first, followed by adding on the asymmetric opening angle. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
106
|
Mon Aug 31 18:32:21 2020 |
Alex M | Daily Update 8/31/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I started working on the slides for our meeting with Wolfgang this Friday. I'll share it via one drive/email/slack tomorrow for comments from everyone.
Over the weekend I finished the asymmetric length run. I still have a couple of fixes to make on plotting scripts, so I'll update those plots tomorrow after making the fixes. I also started a new run today using both an asymmetric length and an asymmetric opening angle. The plots aren't automatically uploading to dropbox anymore because we hit our limit, so I'll have to add them manually. You can see the asymmetric run on dropbox under Machtay_2020_08_27_Asymmetric_Length_Real_Run(SECOND REAL RUN).
|
I'll finish up and share rthe power point with everyone and then fix up the plotting scripts and continue the current run. I also need to try getting onto the UW super computer in advance of the ARA bootcamp on Wednesday/Thursday. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
Futher worked on documentation of the genetic algorithm. Ben is currently doing a big run to test the program. |
Once the big run is finished, we will look at the data and attempt to fix any issues that may arrise. |
|
|
|
98
|
Mon Aug 3 16:39:56 2020 |
Alex M | Daily Update 8/3/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
I fixed the issues I outlined a little while ago that we needed to resolve before starting a real run. I started a real run with 10 individuals for up to 20 generations using the database for a symmetric bicone. The name of the run is Machtay_20200803_Master_Symmetric_Database_Real (whew!). Right now it's on gen 2 and plots are on drop box (see the link at the bottom). |
I'm going to keep the run going and hopefully be most of the way to 10 generations by the end of tomorrow. Julie and I also discussed some potential improvements that can be made to the loop. For one, there's some consolidation and cleaning up that should be done of the main directory. We were also speculating about a way that we could quickly see when AraSim jobs fail and rerun them in real time rather than waiting for all the jobs to finish to realize we need to rerun. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Continued working on the bash script to automate runs for paperclips. |
Finish the bash script and do the runs and start combing through the results. |
| Ben |
|
|
| Ethan |
|
|
Dropbox link: https://www.dropbox.com/home/GP_Antennas/Updates/DailyFitnessScoreImages/Machtay_20200803_Master_Symmetric_Database_Real |
|
|
102
|
Mon Aug 10 16:12:28 2020 |
Alex M | Daily Update 8/10/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Over the weekend I tried making some fixes we discussed. I fixed the roulette algorithm (the symmetric version) so that the random number generator is only set once and then passed to functions. It will now print the values in the generator to files to be plotted. I tried running the loop over the weekend with these fixes but the AraSim jobs kept failing. So today I consolidated the two loops and I started working on a fix for AraSim that will automatically check if an AraSim job fails in real time while the loop is running and resubmit it so that we don't get stuck waiting for the jobs to rerun. |
I'm going to finish the AraSim fix tomorrow and then try starting a run. I'm going to be out on Wednesday and Thursday, so I'll need someone else to keep the run going. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Created a program that combed through all the files, got the average high score of each set of parameters, and saved that information in a master file to quickly use for finding which parameters worked best. |
Create the programs to make the desired graphs we wanted. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
85
|
Thu Jul 9 16:54:27 2020 |
Alex M | Daily Update 7/9/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Finished running the loop for 15 generations. The last plots are on Dropbox under Length_Cutoff_Test_3. There are three things to fix before running this version of the loop again. See below the table.
We started running the next step, which is the database version.
Julie, Ben, Ethan, and I all met to talk about AREA. We started going through the code to understand it, but we're stuck because we don't know if AraSim Lite is actually in the directory or not.
I started rewriting the AREA section.
|
I'll keep the run going, but I might have to stop it and look for bugs if the evolution doesn't track with the previous run. We named this one Data_Base_Test_7_9.
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
Three things to change before doing the next symmetric run (w/o database):
1. Change how the mutation is done. We have at least a partial solution to this--the standard deviations were all 1 due to type errors in c++. We already have a fix for this.
2. The bicone separation distance was too small (0.05 cm). We should increase it to 2-3 cm to represent an actual buildable bicone.
3. We kept the random number seeds in. This isn't a bug, but it's something we need to remember to change before we run this again. |
|
|
84
|
Wed Jul 8 15:09:42 2020 |
Alex Patton | Daily Update 7/8/20 |
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M |
Continued the run from yesterday. Alex P and I applied the changes we made to the bash script for fixing the bugs we found in the simulation number to the database version. Once we get 15 generations of data for this run we'll start testing the database version. Once that works we'll have Eliot and Leo run their version with only 3 genes and if we get the same genes all around we'll be able to merge the asymmetric branch with the database branch and then merge with master. I also fixed up a bug I found in one of the plotting programs that meant it was missing the first individual, it should be good now.
Ryan and I talked a bit about how to apply crossover and mutation to the paperclip. I'm not sure that crossover makes sense, but I think it's possible. He's going to start by doing crossover of the chromosomes and then when that's working he can try crossing over the genes (this is where I'm not sure things make sense, but who knows). Then he can implement mutation in that function.
Julie and I spoke with Ben about AREA and started helping him siftt through all the files. We're going to meet with Ethan tomorrow to catch him up to speed and start reviewing the bash script that runs all of AREA.
|
Finish this run, test the database. I'll meet with Julie, Ben, and Ethan to talk about AREA. Also, Eliot and Leo found a potential issue in the roulette algorithm that I asked them to look at, so we might try making a fix in there and see if that allows our mutations to be more reasonable. Alex and I are going to have to meet with Eliot and Leo soon so we can all work together on merging our branches and resolving merge conflicts.
|
| Alex P |
Made a version of my c++ programs that detect whether an individual matches the database and logs that/writes to the database in a way that allows the number of genes to be a variable so that it can be compatible with new antenna designs. Then set to fix the scripts that used the database to fix the error of the right simulation numbers while continuing the current run so that we can test if the database version gives the same results and then will start running the database version and make sure it is good before we merge. |
Run database test and if it has any differences or bugs from the original, fix those before we merge. Also have Eliot and Leo run thiers but with just three parameters so that we can test and make sure the accuracy between all versions is the same and none have bugs present before we merge. |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
83
|
Tue Jul 7 14:39:12 2020 |
Alex Patton | Daily Update 7/7/20 |
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M |
Found a bug while running the loop, fixed it and starting running again. Added a few things to the paper.
|
Alex and I will keep the loop running tomorrow. We're gonna try to meet with Eliot and Leo to work on implementing the bug fixes we've made into their version of the loop, then merge it with our branch. We need to add it to the database version too, and then after all of that we can merge with the master branch. |
| Alex P |
Now that length constraint is working properly ran a few generations and found a small error with the mutation calculation that allowed for the theta to go below zero, fixed that change and started a run to have a bugless run. |
Continue this run for 15 generations or so and then work on the merging process with Eliot and Leo's branch and then once that is working merge with master once we have confirmed the absence of bugs |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
97
|
Thu Jul 30 15:36:54 2020 |
Alex M | Daily Update 7/30/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Tested the database setting for the symmetric loop. It ran well and gave correct results, so all that's left to test is the asymmetric loop (going to start that momentarily).
Julie and I helped Ben and Ethan set up a git repository for AREA and gave a brief review of using git.
|
Assuming that the asymmetric setting isn't too dificult to get working on the master branch, I'll be able to start collecting real data. I'll need to implement the fixes I outlined last week on Wednesday (see the ELOG post), but those should be quick changes. I'll start by using the database for a symmetric bicone with 10 individuals per generation. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
96
|
Mon Jul 27 16:52:20 2020 |
Alex M | Daily Update 7/27/20 |
| Name |
Update |
Plans for Tomorrow |
| Aelx M |
Leo, Julie, and I merged the two dev branches (Antenna_Database and asymBicone) onto master. I think I resolved all of the conflicts and successfully merged. I'm going to start testing by running each setting (original, database, and asymmetric) through generation 1 to make sure that they give the correct results as the previous test runs and that they have all of the correct files they need to run correctly. Once I'm done with this I'll implement the changes I outlined last week (on the 21st I think) and begin a real run.
I helped Ryan with the paperclip algorithm. We found the error in the roulette algorithm and it now gives results comparable to tournament mode. There is a bug that needs to be fixed (Ryan changed a variable to be taken as an input, but it's giving different results when we input the same values as it had before and neither of us can tell why) but otherwise the script looks much better than last week with the roulette fix.
|
I'm going to finish getting confirmation that the master branch works for each of the settings for the loop. Then I'll be ready to implement the fixes and start a real run, beginning with the symmetric setting with the database. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Issues with low roulette fitness scores have been resolved (there was just an issue with checking the sum compared to the random number) and it has comparable scores to tournament after similar amounts of runs. Then I started work on fine-tuning the mutation function. Early results seem to suggest that having a high mutation rate gets closer to the optimal score faster but lower mutation rates will get closer given longer amounts of time. Finally, Alex concluded that the max fitness score for these paperclips is 9 (or more specifically one less than the number of segments), and some runs after 1000 generations are reaching into the 8.99*** for high scores and averages. So the goal for me this week is to try to reach that point faster (hopefully within 100 gen). |
Continue fine-tuning parts of paperclips to find optimal methods. If I reach that, write a bash script to autorun tests. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
95
|
Fri Jul 24 16:25:06 2020 |
Alex M | Daily Update 7/24/20 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Ryan |
|
|
| Evelyn |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
94
|
Thu Jul 23 16:44:05 2020 |
Alex M | Daily Update 7/23/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Leo and I continued working on the roulette algorithm for the asymmetric bicone to try to get the evolved generations to match the original and database ones. We went through the mutation part of the algorithm and made sure we were rrunning all of the random number sequences in the correct order (for example, we need to make sure that if we define variables A and B from a random number generator, that in both cases we define A first, then B). We also had to make sure that we were using these random sequences the same number of times (for example, we don't want to define A once in the original/database versions and twice in the asymmetric version when the asymmetric version is set to evolve symmetric antennas--this puts the random number generator "off sequence" relative to how it is in the original/database version). We tested this with the gen 0 DNA and effective volumes (which matched with the results from the original/database runs) to evolve generation 1 and got it to match. I'm currently running the loop to test this over multiple generations.
Julie and I met with Ben to talk about AREA. Yesterday, Ben was able to generate the 0th generation of individuals and then run AraSim from a job to get effective volumes. Today, we focused on editing the scripts to work for OSC job submissions (he wrote the one he used yesterday; we modified the main script today and will continue to do so). We edited it so that we could submit the main script as a job which then submits other jobs for the arasim portion and waits for those jobs to finish before continuing on (since it needs those effective volumes for its fitness score). We're going to keep incrementally fixing the main script so that it can run over multiple generations as a job submission without errors.
|
I have a doctor's appointment in the morning, but maybe Leo can continue my current run. Once it's done, optimally we would work with Julie to get everything merged so that we can start a real run (we also have to implement the changes I outlined in the ELOG post a couple days ago before we do a real run/merge--most of those are pretty simple).
Julie and I will take a look at the paperclip script Ryan's been writing to see if we can help him figure out why the roulette algorithm is giving such worse fitness scores than the tournament algorithm (~ fact of 2 worse).
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
Alex covered it above for the most part. I just worked with him today trying to correct roulette so we get the same DNA for gen 1. Just to reiterate, the last step that was hanging us up was making sure the new GA used the random number generator the same amount of times and in the same order as the old GA. After that, Alex started another run to make sure everything is running smoothly. |
Tomorrow, Alex and I will make sure the results for the first few generations match. Then, Alex, Julie and I will move on to merging. |
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
92
|
Tue Jul 21 17:01:58 2020 |
Alex M | Daily Update 7/21/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I replaced AraSim on Eliot's user with the one we have on the shared space since I wasn't able to compile his yesterday. It compiled correctly and I ran the loop but got the same results from last week (effective volumes that differed from the ones we got in the original and database test runs). I found one potential error in the seed we were passing to AraSim from the loop, so I'm currently running with that fix. I'm hopeful, but not convinved it will fix things because the setup.txt files on each version of AraSim looked the same before I made this fix. Regardless of whether or not it fixes the problem, it was a problem so it was important to run with that fix. This also gives me another thing to add to the list of changes to make before doing a real run. You can find the list below. |
I'll continue this run for 4-5 generations if it looks like it's working just to confirm that we're getting agreement with the previous runs. If it agrees, I can turn my attention to either merging or doing a real run. I can do a real run without merging, but it depends on if Amy wants me to merge first or not. Eliot gave me his account info for OSC and github so I should be able to merge correctly (hopefully I can meet with Julie over this so we can make sure we don't have a problem) but it might take me some time to resolve the conflicts. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Worked on rewriting certain parts of the paperclips code so that it runs better. |
Fix the bugs in the code. |
| Ben |
|
|
| Ethan |
|
|
Here's the list of changes to make when doing a real run (undo some of these when doing a test run):
1. Change the separation distance from 0.05 cm to 2-3 cm.
2. Remove the seed in AraSim by changing the setup.txt file so that Random_Mode = 1 instead of 0.
3. Add noise to AraSim (unless Amy says not to?) by changing the setup.txt file so that Trig_Analysis_Mode = 0 (or whatever the setting is--see the comments).
4. Make sure all of the frequency lists are the same! I think this has largely been resolved, but basically there was a time when the frequencies we were using differed from those used by AraSim for the actual bicone. We changed our lists to agree with the AraSim one, but I think there could be a few lists floaitng around (potentially just commented out) that I'm going to want to take our or correct.
5. Remove the seeds from the roulette algortihm. To do this, search for where "seed" is used and comment out the one that has 1 as an argument and comment in the one that uses time(0). Also, do the same for srand().
6. Fix the exponent in the calculation of the standard deviation of the genes in the roulette algorithm. Right now it has an integer division error, so the exponent becomes 0 leading to all the sigmas being 1. This is an easy fix--I'll just change 1/2 to 1.0/2.0.
I haven't made these changes yet because we are testing and want to confirm that we get the same results from the asymmetric version with the original and database versions. Since those didn't have these fixes, I haven't put them into the asymmetric version yet either. |
|
|
93
|
Wed Jul 22 16:00:18 2020 |
Alex M | Daily Update 7/21/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Leo and I worked on the roulette algorithm to figure out why the new individuals being formed are different in the asymmetric bicone version of the loop than the original and database versions. We made some fixes, but we came across a complicated problem. Essentially, we think that the difference is coming down to the different number of times the random number generator is being called. Oddly, we can get the lengths that are generated to be the same. But getting all of the genes to be the same would require substantial reworking of the structure of the roulette algorithm and how to do it isn't obvious. We can keep trying to figure it out, but the restructuring was important to allow the mutations to occur on either chromosome (vs in the original and database versions, where we just have one chromosome to mutate, here we need to allow for more mutations since we have up to 6 genes). In principle though, I feel like this is possible, just difficult and requires substantial rewriting. |
I'll keep looking at the roulette algorithm to see how we can get the same new individuals as the previous two runs. Julie and I were also planning on meeting with Ben about AREA to better understand how to run it properly, |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
I fixed the bugs that I was having yesterday and the algorithm should run more properly. I also added functionality for sending certain parts per ten individuals to either tournament or roulette. However, neither function is getting "optimized" results by 100 gens (though the tournament is significantly closer than roulette in isolation) with the highest fitness score I have been able to get was ~8.65. But, I also do not know what the theoretical max fitness score, given the parameters, would be. |
Find out what an optimized fitness score's value would be so I know how close I am to achieving that. Run preliminary tests to see what combinations work best for now. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
91
|
Mon Jul 20 17:01:35 2020 |
Alex M | Daily Update 7/20/20 |
| Name |
Update |
Plans for tomorrow |
| Alex M |
The arasim jobs I submitted over the weekend kept failing. I've been looking for the cause but I can't pinpoint it right now. The issue arose on Friday when I overwrote the version of Report.cc on Eliot's user with the one we have on the databse branch (but I don't think there were realy differences aside from comments). Before I had done this, we were getting effective volumes that differed from the ones we got on the database branch. Now, the araSim jobs are just failing after 5 seconds of running. And now I'm getting an error when I try to recompile AraSim (I might need Eliot to do this?).
I tried getting an interactive job so I could confirm that things are still working on the database branch, but I couldn't get it because it was blocked. I think that was because we had too many concurrent jobs running, which has happened a couple times over the past week-week and a half or so.
|
I need to track down why these AraSim jobs are failing. Besides maybe needing Eliot to make AraSim, one difference I noticed is that it appears we are sourcing Lauren's version of ANITA build tools rather than the one on the shared space. But I couldn't test this because I need to recompile AraSim. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
80
|
Thu Jul 2 15:03:39 2020 |
Alex Patton | Daily Update 7/2/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Alex to change the roulette algorithm to implement a cut on the minimum length for the Antennas. Amy showed that we don't want to generate antennas which are shorter than 37.5 cm in length (for each cone, so a total of 75 cm all together) because anything shorter than that won't be reliably simulated in XF for our frequency range. To do this, we made the roulette algorithm reselect individuals in the 0th generation if they were below this length. When it's making new individuals for the next generation, we made it so that the algorithm doesn't apply mutations that would make the antenna smaller than the minimum length (but we may revisit this with other methods, like letting the individual die if its below that length).
Spoke to Ben about AREA. He can run it, but there are some errors, and it looks like the job was killed when we tried submitting it to the queue. We'll have to see if we can track down some of the errors.
|
Alex and I will try to run the loop using our changes to the roulette algorithm and see what the new individuals look like and if the effective volumes are reasonable. |
| Alex P |
Worked on converting the c++ programs I made for database to work with a variable amount of genes rather than just LRT. After that Amy met with Alex and me and discussed the proper length to limit the antennas to and then worked with Alex to put a clamp within the roulette algorithm on both the initial generation and the generation of lengths through mutation. Then deciding to just cut the individuals that have too low lengths and discussing how that would be implemented. |
Will continue to implement and test the changes to clamping the length by eliminating low length individuals and once that is sorted start working on being able to make our changes work on other branches too so we can start to merge. |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
90
|
Fri Jul 17 18:09:16 2020 |
Alex M | Daily Update 7/17/20 |
| Name |
Update |
Plans for Monday |
| Alex M |
We may have found the error in the asymmetric bicone. It appears there may have been two problems. First, the setup for the simulation_PEC.xmacro script in the asymmetric version appears to have been slightly different in that the positions of the cones, relative to the positions of the coordinate systems, are different from in the other versions of the loop. This was giving different uan and gain files than the previous runs did. We fixed this, but it revealed another potential issue: in order to evolve the separation distance, we need to make it so that the coordinate systems use the separation genes. This wasn't implemented in the asymmetric version, so there would have always been a disparity in the position of the cones relative to their coordinate systems when compared to the database version of the simulation_PEC.xmacro. I have a work around for when we don't evolve the separation distance, but if we want to evolve it we need to change this script.
Second, the version of Report.cc in AraSim on Eliot's user differed from the one in the database version. The other files agreed, so I copied our version into his directory and started a run with that. It's running now and should take ~1 hour before I can see if this has fully fixed the issue.
|
I might be out on Monday, but my main goal for next week is to see if the fixes we implemented today resolve the differences in effective volumes and evolved parameters. Once we have them in agreement, we can start a real run on the database version of the loop to get symmetric data for the paper. While that happens, we'll need to try fixing the asymmetric simulation_PEC script so that it can use the separation distances properly. Then we can test that again using those separation distance variables (I used a hardcoded value) to make sure we get correct results. After that we can merge the asymmetric branch with the database branch and then the database branch with master and get data for the asymmetric version of the loop. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
89
|
Thu Jul 16 15:18:32 2020 |
Alex Patton | Daily Update 7/16/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
(See below the table for a review of the past couple of days since ELOG has been down.)
We may have found what was causing the error in Eliot and Leo's version. They were getting 0 effective volumes. I edited their output macroskeletons to use pro grid (I figured it was the default setting, but it looks like not). This seems to have fixed the issue with their .uan files having all 0s and 180s for the phase and -300 gain. However, it wasn't writing to the vEffective.csv and fitnessScores.csv files correctly. We realized they may have needed to recompile. We did that and ran a preliminary tset, where we got some promising results. The effective volumes and fitness scores seem to have been read out and plotted correctly, but they didn't appear to match the previous two runs Alex P and I did. That might be because we were messing around with the save state sloppily. Currently, Eliot is rerunning from the beginning with the intention of seeing if it will go all the way through generation 0 smoothly. From there, we'll need to compare the effective volumes and fitness scores with the previous two runs Alex P and I did (although I can confirm that the generation 0 DNA matches). Then we'll need to let it run for a few more generations to make sure it evolves the same individuals those two previous runs evolved.
|
If it looks like these fixes worked and the effective volumes are being made correctly and the same individuals are evolved as in the previous two runs Alex P and I did, then we'll be ready to start merging. The first step will be to fix a merge conflict on the asymmetric bicone branch. Then, we'll have to carefully merge and resolve merge conflicts. The order should be that the asymmetric bicone branch merges onto the database branch, which will then merge onto the master branch. Once that's done, we'll be able to do a proper run.
|
| Alex P |
Worked with Alex, Eliot, and Leo to fix an error on Eliot and Leo's branch since we got both the normal and database version working on the Alex's branch. Had some problems with 0 vEffectives and saw that XF was giving 0 gains so searched through for a while to see what was different and had to enable PrOGrid, and then that made the vEffectives nonzero but it still seemed like they might have been too high. |
Get Eliot/Leo's branch working hopefully have test run overnight that shows that and then merge tomorrow is the plan. |
| Eliot |
|
|
| Leo |
Like both Alexs said, Eliot and I worked with them to get our version to get the same results as their version. The main problem we worked on were the 0 vEffectives and fitness scores. We realized that the fitness scores and vEffectives were printing to the terminal, but not to the plots or .csv files. We check the fitness function called in the loop and it turned out we compliled to a wrong version. After recompiling the fitness score correctly Eliot started a new run and it is currently running over night. |
Tomorrow, we will take a look at Eliot's run to see how it did and find any other areas that may cause for different results between our's and the Alexs' runs. |
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
Worked with Ben on running AraSim and fixing errors it would output |
|
Review of the past two days (wrt the bicone loop) since ELOG has been down:
Alex P and I were able to fix errors in the database version of the loop. It now evolves the same individuals as the original version (when using seeds). We want to test the asymmetric version, but we ran into some errors. First, we needed to add in the cutoff length and the change in Detector.cc to set negative gains equal to 0. Once we did that, we tried running but found some more errors. Specifically, the effective volumes coming from AraSim were all 0. We may have fixed the error today, but we're waiting for the run to finish the 0th generation to be able to tell.
|
|
|
Draft
|
Mon Jul 13 15:34:40 2020 |
Alex M | Daily Update 7/13/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
Alex and I continued running the loop for the database test. We ran into an error over the weekend that was affecting generations after gen 0. Alex fixed it this morning and continued the run.
The goal of this run is to test the database. We are comparing it to the previous run we did last week to make sure that we get the same individuals. We're using the same parameters as the previous run: NPOP of 8, separation distance of 0.05 cm, and the same roulette algorithm (AKA the same seeds). In principle we should generate the same DNA for the individuals.
Starting in generation 2, we have a divergence--one of the individuals is different from generation 2 of the previous run. We think this is because there is noise in AraSim (I don't think we can turn the noise off when we're using the effective volume). This makes the probabilities of choosing each individual different and allows different ones to be chosen even though the random number being selected is the same. Oveall it looks ok and we're going to keep the run going to see if it converges to the same individuals as the previous run after a few more generations.
I also wrote a bit in the paper. I took a look at the paper Amy sent on the effective volumes to try to understand it better. I think I understand it, but I'm trying to see how the equation we used in AREA in the proceeding is related (what the weights represent).
|
We'll try finishing up this run and doing Eliot and Leo's run. If we can get results fast, we can start merging. Our intention is to start a real run on Wednesday. That would likely take ~1.5 days. |
| Alex P |
I made a fix to the database over the weekend when I noticed it was incorrectly counting the array indexes for counting failures. And then I found an error in our output.xmacro generation when making one for the database with the simulation number fix and that xmacro wasn't using the right bounds. We got the database version working through multiple generations. On the second generation the individuals matched exactly to our previous run except one individual. Looking at the fitness scores of the previous run, the parent individual in the previous generation had a fitness score of .18 compared to 4's of other parents, so it's reasonable that it wouldn't get picked from another run of the algorithm. While the GA is seeded so that it makes the same changes when it picks the same parent individuals, the fitness scores vary slightly between these two runs which alter how the parent individuals get picked especially when it comes to smaller fitness scores like this.
Also fixed an issue on Eliot and Leo's copy of AraSim, the extrapolation issue we found earlier hadn't been implemented on their branch so to make sure all the tests before merging were accurate we made that change.
The name of this run is Data_Base_Test_7_10.
The name of the previous run we are comparing against is Length_Cutoff_Test_3.
|
Continue running this overnight to get results and hopefully start merging tomorrow or at least start the process of setting up for the merge and finishing up any of these runs. |
| Eliot |
Began a final test run with constant separation to ensure 2 chromosome loop is working before we merge. Stopped during gen 0 to implement progrid in arasim. Thanks Alex M! Continued the run from just before arasim. Had very slow progress due to bad wifi connection. I kept getting kicked off OSC! So far everything looks good but wifi made for a shotty test at best. Name is FERSTL_20200713_PREMERGE_TEST. Lastly, commited one last time before we merge. Had some issues with that as always but believe I have it worked out. |
Will try to run a couple more gens on that run and begin merging with the Alexs. |
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Created a temporary mutation function to test to see if the convergence problem from Friday would be resolved. For the most part, it did resolve the problem and the function has not converged onto a single paperclip and the fitness scored being produced are higher than before. However, I noticed that in test runs with on average greater than ten individuals (usually) the individuals being chosen are the same every run regardless of fitness scores and it seems to be causing the scores to stall out. for example in a run of 100 generations the 90th-100th generations would only pick the 0th and 1st individual for all ten chosen. Even when seeding the random number generator, this appears to be happening and I am not sure why. Some runs with higher chances of mutations seemed to have this effect happen later than ones with low mutation chance but so far this just seems like a correlation and not causation. |
Fix the aforementioned issue. Then, rework the mutation function to more closely model the one in bicone's roulette algorithm. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
Draft
|
Mon Jul 13 15:35:06 2020 |
Alex Patton | Daily Update 7/13/20 |
| Name |
Update |
Plans for Tomorrow |
| Alex M |
|
|
| Alex P |
The name of this run is Data_Base_Test_7_10.
The name of the previous run we are comparing against is Length_Cutoff_Test_3.
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
86
|
Fri Jul 10 16:35:26 2020 |
Alex M | Daily Update 7/10/20 |
| Name |
Update |
Plans for Monday |
| Alex M |
Yesterday we started a run of the database version of the loop. The goal was to use it to replicate the results from the run we finished yesterday using the same parameters (NPOP=8, SEEDS=10, bicone separation = 0.05 cm). We initially ran into problems with the databse which we fixed this morning. We successfully completed the 0th generation, but then the .uan files for the next generation weren't created properly. Alex and I will try to resolve this. Amy suggested we only need to run 6-7 generations to confirm that we get the same results as from the previous run. The 0th generation agreed with the 0th generation of the previous run (in terms of DNA; the effective volume/fitness score plots looked similar, but there is slight room for noise in AraSim so the exact values are slightly different; but overall they look very similar). I also confirmed that the generation DNA is the same in generation 1 of the current database run as in the previous run.
The name of this run is Data_Base_Test_7_10.
The name of the previous run we are comparing against is Length_Cutoff_Test_3.
|
The top priority right now needs to be with fixing the error in the database version that is causing the uan files to be made incorrectly. Once that is fixed, we can continue the run and confirm that we get the same results After that, we'll have Eliot and Leo run their version using the same parameters to confirm that they get the same results as the previous two runs. From there, we'll merge the branches and begin a real run.
The first real run will use NPOP=8, 3 dimensions (LRT), a separation distance of 3 cm, and will run for up to 15 generations. We'll also fix the issue of the mutation standard deviation (see yesterday's update for the three differences between our current runs and the future real runs).
Our goal is to get as much real data as possible next week. I'm confident that we can have at least one with 3 dimensions (LRT) done by next Friday (we'll shoot for 15 generations, but if it seems to converge between 10-15 we can stop it).
If we can finish that run, we can start another run using 4 dimensions, where we add an option to make the bicone assymetric in length (L1L2RT). These are the runs we're hoping to use in the paper.
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
Today we looked at the results of our last run. This run consisted of 8 individuals and was set at 20 generations, but we just wanted to see how far it would evolve. We noticed that the error came from LRTS.py when we need to reshape the arrays from gensData.py in order to plot them. The error said that the array couldn't be reshaped because the new array's dimensions were too small. After digging around and rerunning with past save states, we were thinking that the issue wasn't with the code, but was actually an issue with generation the loop said it was on. For instance if we were on gen 5, LRTS thought we were on 4. Eliot was pretty sure this was because we had been interupted during part of our run, so Leo gave him the benefit of the doubt and decided to run again. This time we only ran 3 individuals up to the 1st generation, just to make sure the plots were working. Leo was running, making sure not to have any interuptions, and indeed all the plotting softwares worked. So, to the best of our knowledge our version of the loop is ready to go. |
Next week we are just waiting for the Alexs to finish up with their testing so we can start merging. We didn't have any plans to work on outside of that. |
| Evelyn |
|
|
| Ryan |
Finished the crossover function for Paperclips and ran a few tests for the roulette + crossover. Tests seem to show that the paperclips quickly (between 3-5 generations ) converge to become the same paperclip and fitness scores (determined by z-curl) dropped off after the second generation. This is most likely due to the fact that currently there is no form of gene mutations written into the crossover function for repopulation and so the angles the segments can have are locked from the first selection. However, I am not 100% certain on this and there could just be a hidden error somewhere as at times in some (not all ) of the tests I did there were times where the roulette function passed many of the same paperclip to the next generation, therefore decreasing the size of the gene pool. I need to do further tests to be certain of one or the other but otherwise, everything seems to be working as intended. |
The next step I plan on taking is creating a mutation function in effort to make the gene pool larger. It is likely that I will look at the bicone's roulette program to model this mutation but I am considering writing a simple one first just to run tests on the effect on the diversity. After this step is complete, I will move on to creating a function that is able to send certain amounts of the population to the tournament and the rest to roulette to see what mix gives the best results. I also as a pre-step to this may take the existing algorithms in paperclips and re-write them as functions rather than loops in main to make the passing of different parts easier and faster. |
| Ben |
|
|
| Ethan |
Continued documenting AREA project in google doc. Worked with Ben on getting AraSim running and fixing errors. |
Meeting with Ben to continue on AREA project. We may try to get AraSim run the children in parallel to make it go faster as it takes a very large amount of time currently. |
|
|
|
79
|
Wed Jul 1 16:18:35 2020 |
Alex M | Daily Update 7/1/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked to make the frequency plots up to ~2.5 GHz. Amy said we should put a cut off for the length of the antennas based on the minimum frequency, but right now I'm not sure how to find what that minimum length should be based on the frequency (we should be using the lowest frequency for this calculation though, since it corresponds to the highest minimum length). |
I'll keep looking at how to figure out the minimum length and implement it into the loop. |
| Alex P |
|
|
| Eliot |
Implemented the asymmetric radius and separation parameter throughout the rest of the loop. Began a first test. |
Will check on and fix any errors and either tonight or tomorrow do a run for Alex to see about length convergence. |
| Leo |
|
|
| Evelyn |
Continued working on code academy, got about 3/4 of the way done |
Try to finish the code academy class before my free trial runs out! |
| Ryan |
Fixed the issues in the roulette function from Monday and integrated it into the rest of the code. seems to be running fine so far. |
Next step is to code in a user input for how many of ten individuals get selected by tournament vs roulette. Once this is complete I will try to run some tests and probably try to have the results output to a file to save results for comparisons. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
65
|
Tue Jun 9 15:00:09 2020 |
Alex Machtay | Daily Update 6/9/20 |
| Name |
Today's Update |
Plams for Tomorrow |
| Alex M |
Alex P and I kept working on finding differences in AraSim between antennas. We still haven't found any and we've checked: electric field, maximum peak voltage, effective height, threshold (factor and offset), and the fft (and maybe more I'm forgetting). I'm not sure where to go from here in investigating AraSim.
I tried running the loop with a smaller grid size to see if we could get some more comparisons, but I ran into errors that I need to resolve.
Julie and I resolved some comments on the paper.
|
|
| Alex P |
Went through more of AraSim after more of Amy's comments. Found V_forfft and they were the same between individuals and then looked for other information to compare but what is interesting is that with the smaller grid spacing on the smaller antennas we have two generations with the exact same dimensions and the smaller grid spacing had lower vEff across the board, such as the larger grid spacing on small antennas had 4 individuals with a vEffective above 5 but the small grid spacing didn't have any that got above 5. So it is possible that the zero vEffective for the small antennas wasn't an error but that the larger fitness scores were the error. |
Plan is to continue to look into how grid spacing affects the vEffectives especially of smaller antennas and also make sure the grid spacing we were using for the larger antenna is appropriate and then look into the automatic grid spacing whether we can do it within XF or by having it automatically update the Xmacro based off the lengths |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
78
|
Tue Jun 30 17:12:32 2020 |
Alex M | Daily Update 6/30/20 |
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M |
Continued looking in XF for why the frequency where two antennas separated by a small amount give the same gain. I made a script which plots the gain at each frequency for a chosen number of individuals at a specific polar angle and used it on two antennas, one with a separation of 0.01 cm and one with a separation of 0.005 cm. The plots look similar, but there is some difference--see the genstudents slack. |
I'll continue to work on figuring out the frequency where these antennas start to give the same gain. Our ultimate goal is to find a minimum length for the antennas to cut off at so that we don't evolve antennas that are too small. |
| Alex P |
|
|
| Eliot |
Made little progress on the separation gene throughout the loop, however we know where all we need to make changes(I think). |
Should be able to make significant strides in regard to separation throughout the loop(ie plotting, fitness function, etc). Hopefully can also begin testing and debugging. |
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
77
|
Mon Jun 29 15:57:41 2020 |
Alex Patton | Daily Update 6/29/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Alex P to look at the different gain patterns for antennas with different separation distances. We found that there wasn't a clear frequency where they started to agree--looking at the average frequency gain plots we sent on slack, they look very different. We're trying to resimulate them with some higher frequencies to see the pattern more clearly and find where they start to agree. |
I should be able to finish making those frequency plots in the morning. I think I'm going to try spending some time with Karoo tomorrow and see what Julie's been able to figure out. |
| Alex P |
Looking at differences between small antennas and how the spacing affected their gains to see where differences occurred and saw at 350 MHz is last one where bigger spacing is better and after that it seems like the smaller spacing is better at 366.7 MHz and above. We checked and it actually flips back and forth between which is better and couldn't find a real pattern there. Switched to make an average gain plot for each frequency to see a pattern in the average gain since the 3D graphs use color and sometimes it can be hard to tell by how much the differences are. |
Continue to work with the spacing to find which frequencies cause problems for antennas according to their sizes. |
| Eliot |
Implemented the option of an asymmetric radius within the GA. Now, our new gene (separation) is the only gene without the possible asymmetry. This is because an asymmetric separation wouldn't do anything as the separation in XF(ie its offset from the origin) does not constitue a physical significance in AraSim. |
Leo and I will begin to implement the separation gene throughout the remainder of the loop. |
| Leo |
|
|
| Ryan |
Continued working on the roulette function for paperclips1.0.1.cpp. Ran into some issues with returning an array. |
Debug the roulette function, maybe test if I get everything working and sent to the correct places. |
| Evelyn |
|
|
| Ben |
|
|
| Ethan |
|
|
|
|
|
75
|
Thu Jun 25 15:22:50 2020 |
Alex Patton | Daily Update 6/25/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Alex P on figuring out why we keep evolving towards small antennas. We think it's because the initial angle is so large, and the standard deviation of makes it more difficult to bring the antenna within the confines of the hole by mutating the angle than by mutating the the length. So we started a run with a much lower angle (pi/24 instead of pi/12) to see if that yields antennas which are longer with better effective volumes.
I also edited some more of the paper and added in a few citations, but I'd like to make sure that those citations are written correctly (for example, I couldn't find the eprint for all of them).
|
I'll keep this run going tomorrow and monitor it while I write some more in the paper. Now that we have a better understanding of the xmacros and can have debugged a lot, we can probably start a real run soon and I can write some more details in the PAEA section of the paper. Also, since I'm better acquainted with XF, I think I'll be able to get images of the bicones from there to put in the paper. |
| Alex P |
Running and ran into some errors with XF solver. Found a fix to it but came into a possible oversight. So I think one reason why we keep getting this trend towards lower antennas is that the initial thetas from the 0th generation in the GA are too large and that makes it so all the longer lengths end up making too wide antennas which get penalized and hence why it tends toward small because while we do have mutation to make different thetas it only varies by so much and I think the opening angles are just too large to start in the distribution. This doesn't fully solve why the small antenna tend to give higher results than expected but it still is a problem because it's causing all the larger gen 0 antenna to get a zero fitness score because they are almost always too large. Currently if all the mean values in the distribution were used to make gen 0, the outer radius would be way bigger than the hole by a factor of 2. |
Started run with change to the GA to have a lower theta as the mean in the distribution. Will see if this starts to favor the larger antennas. Possibly look more into why that XF solver error was being caused and effects of changing the spacing between cones on the bicone. |
| Eliot |
|
|
| Leo |
Today we started work on adding seperation as a 4th gene. We updated the GA and tested it in a mini bash successfully. We then moved onto editing the Xmacros scripts and tested using a GUI. |
Tomorrow we plan to move onto the fitness funciton and plotting scripts. |
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
133
|
Wed Jun 23 16:34:10 2021 |
Ethan Fahimi | Daily Update 6/23/2021 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Lydon |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
With Alex M's help, managed to get AREA working, plotted results. |
The results look relatively flat, possibly the GA is unoptimized, may need work. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
|
74
|
Tue Jun 23 14:37:07 2020 |
Alex Patton | Daily Update 6/23/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Aelx P on running the loop with decreasing grid spacing. I also worked in the XF GUI some more and figured out how to make 3D gain plots (per Cade's suggestion) and tried comparing different grid sizes for a single antenna with proGrid on.
|
Alex P and I are gonna meet with Cade tomorrow to talk about XF some more so we can figure out where progrid is having an impact and decide on what the best idea is for making sure we get the correct effective volumes out of AraSim. |
| Alex P |
Continued today running the loop and while we had the next level of smaller grid spacing running, tested how to get the PrOGrid working and managed to get a script that would work with it. We got rid of the grid size variable but kept cell size as a constant. Although there should also be an option to make cell size automatically set via PrOGrid and there are other options that you can enable PrOGrid to handle as well so we may need to mess with more to find which are valuable. |
Figure out how PrOGrid affects vEff compared to the variety of Grid sizes we've tested and also look at this for larger antennas too. Possibly tweek more settings too |
| Eliot |
|
|
| Leo |
After testing the GA outside the loop with a initial generation, it was still giving the same "stoped" error. We thought it might be getting stuck in an infinite loop somewhere and decided to add a few flags to get a better idea of where. We found in was during a section that checks that Tourney_pop+Roulette_pop=NPOP but this was never achieved so it was getting stuck. This is because we had 5 individuals and had tournement proportion set to .5 with an NPOP of 5, so we think the GA didn't know what to do with 2.5 individuals in each. To fix this we changed tournement proportion to .4 so we would get 2 and 3 individuals in each. We then did a run and noticed that tournement was having some issues. So, we ran with tournement proportion at 0 and are going to fix tournement later. We ran, but our second generation was interupted in XF by someone else's run. We waited a few hours for people to finish and ran again at night. |
Check on results of last run to see what other errors need fixing. Also, check on tournement and try to fix those errors. |
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
73
|
Mon Jun 22 13:52:32 2020 |
Alex Patton | Daily Update 6/22/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Alex to run XF with smaller grid spacing. We're down to 0.001 cm, but below that we were getting long times on the XF GUI that made it seem like it was hanging. I also investigated XF from a Pitzer desktop to see how to use the progrid setting. I can get the scripts to load--I checked that the original one is indeed making a bicone and that Eliot and Leo's makes an asymmetric bicone.
Helped Ryan some more with the paper clip. We're taking it step by step, so the next thing he'll want to do is make sure he can get the function for the roulette algorithm to be called correctly in the paperclip script and have the correct information passed to it (like the fitness scores).
|
I'm gonna continue investigating XF from the pitzer desktop to see how the progrid works. When Alex and I finish this generation of the loop, we'll try putting the progrid in an see what the results from XF are to compare it to the results frmo previous generations when we explicitly set the grid size. |
| Alex P |
Along with continuing runs of XF to find optimal grid spacing also looked through the HTML files in the XF directory to find the scripting documentation. Found where the PrOGrid options were along with the other features PrOGrid enables. Currently we have other variables that use our Grid Size as an input so we would have to find a way to replace those to work with what PrOGrid generates. |
Continue to check on status of grid size for our current runs and its affects, we might have found a cut off point so we can go back and find the optimal spacing for small sizes before swtiching to tests with large sizes. And possibly start experimenting with PrOGrid since we can compare the output it gives to an array of different grid spacings to see if it would need any tweeks. |
| Eliot |
Continued debugging the Loop to work with 2 chromosomes ie 5 parameters. Ran inside and outside the loop trying to find the error. Finally, we placed a number of flags throughout and determined the error to be within the GA. It is inside the roulette function. We will edit this tomorrow. |
Edited the roulette selection as needed. There is also an error in tournament, but for now we turned it off and are focused on testing the updated roulette. |
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Debugged the Roulette algorithm and have it running. But, I need to check the Gaussians against the paperclips tournament to refine it and make sure I can get a fitness score for the paperclips. |
Get fitness scores for the paperclips and begin to compare to tournament. |
|
|
|
72
|
Fri Jun 19 16:27:53 2020 |
Alex Machtay | Daily Update 6/19/20 |
| Name |
Today's Update |
Plans for Monday |
| Alex M |
Ran the loop with the same individuals in each "generation" to test how decreasing the grid size would affect the effective volume. Using individuals with lengths of 5cm, I startde with a grid spacing of 0.01 cm and kept cutting down by a factor of 2. I'm down to 0.00125 cm right now. Once I find where the effective volumes stop decreasing, I'll do a binary search for what the optimal grid spacing is for that size. |
I'll try doing the same thing that I'm doing for small antennas for large ones to see what the optimal grid size is there, but I think ultimately we're going to want to compare to the progrid to see if XF optimizes better. Amy came up with an idea for finding a function for what the grid spacing should be at a given length, so we'll try to compare that to the progrid. Meaning I'll spend Monday looking through more XF scripting while the large antennas run. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
71
|
Thu Jun 18 14:33:32 2020 |
Alex Patton | Daily Update 6/18/20 |
| Name |
Update for Today |
Plans for tomorrow |
| Alex M |
Alex P and I implemented the changes we discussed with Amy to Detector.cc. We're currently running a test to see how the grid spacing affects the gains from XF. We started with small antennas using a grid spacing of 0.01 and we're cutting down by factors of 2 each generation (until we see that the gains stop changing, then we'll do something of a binary search between the last two grid sizes to search for the largest reasonable size).
I also played around with the XF scripting more. My goal is to try to be able to edit the scripts we use to do help us check our results, like save images and load models for the antennas. I can currently run our macro script in XF by loading it in with 1 individual (using a generationDNA.csv file I moved to my user space). I can view and play with the model and it saves an image of it when I run the script (though the image doesn't give much detail). Hopefully this will lead to better understanding XF and being able to check our work and use the progrid option.
|
I'll continue with the grid space test tomorrow morning and try to play with XF a bit, but I haven't gotten to address many of Amy's comments on the paper from last week so I'll probably prioritize that while the loop is running the AraSim jobs. |
| Alex P |
After asking about the change to replacing negative gains with 0 in the lower frequencies, we implemented that and ran a few individuals through that. We started a normal large antenna run with that change but stopped in order to test the affect of grid spacing on small antennas. We ran a .01 grid spacing and then .005 grid spacing on the same individuals and will continue so on until we see no change in the results. Specifically checking the affects at the 300 MHz frequency but also overall results. |
If this works we will find the proper grid spacing for an antenna's size and the next step will be to implement a feature to properly scale that with the antennas. If we still encounter the smaller antennas giving an abnormally high vEffective we will probably have to continue looking into what could be causing that. I won't be online most of tomorrow, I might check in at a few points but Alex M will handle running the tests instead. |
| Eliot |
|
|
| Leo |
Did a run with 5 individ and 5 generations. It successful ran through the loop 5 times, but didn't seem to evolve. In particular, "GenerationDNA" wasn't changing throughout the 5 generations. |
Tomorrow, we plan to investigate where this error is coming from within the loop. As of now, we aren't sure whether this is part of the loop or GA. Also we are getting an error message about the file "AraOut_ActualBicone" but doesn't seem to stop the loop, so we want to check in about this as well. |
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
70
|
Wed Jun 17 17:21:16 2020 |
Alex Machtay | Daily Update 6/17/20 |
| Name |
Update for Today |
Plans for tomorrow |
| Alex M |
Worked more with Alex P on AraSim. We've found the cause of the low effective volumes. The function GainToHeight in Report.cc uses another function called GetGain_1D_OutZero in Detector.cc. The problem is that for some small individuals, we get that the output from GetGain_1D_OutZero is negative, which leads to -nan values from GainToHeight because it takes the square root of the negative values from GetGain_1D_OutZero. We aren't quite sure *why* we're getting those negative values (we know which variables and operations are giving them, but not why they're so different for some small individuals), but we think a solution is to set any negative output from GetGain_1D_OutZero to just be 0, since they're all lower than the lowest frequency in the band of interest anyway. |
We can implement the change to fix these effective volumes. I'm still concerned that we're getting high effective volumes from this, when I would have expected the resolution to have decreased effective volumes for all of the small antennas. We should also be testing this with the actual Bicone to see how it changes that effective volume. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Possibly made progress with the roulette algorithm for paperclips. However I will not know for sure until I fix the syntax errors in the code ( which seems to be largely just me mistyping something up to this point). |
fix the errors in roulette.cpp and make sure the output is desired. Fingers crossed. |
|
|
|
69
|
Tue Jun 16 16:10:52 2020 |
Alex M | Daily Update 6/16/20 |
| Name |
Update for today |
Plans for Tomorrow |
| Alex M |
Continued working to find the AraSim error. We think we're very close to finding where the difference between the two individuals (one with nonzero effective volume, one with 0) is arising. We've pinned it down to a series of if statements around line 952 of Report.cc. We should be able to piece it apart tomorrow. |
Continue working on AraSim to find the error we think it in that if statement. |
| Alex P |
|
|
| Eliot |
Tried to run a couple tests of asymmetric bicone. Finished up on the git codecademy. Getting close to being ready for merge. |
Test runs, meeting, hopefully develop new plans. |
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
68
|
Mon Jun 15 15:52:45 2020 |
Alex Patton | Daily Update 6/15/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Worked with Alex P on looking for the problem in AraSim. We looked in Report.cc where Amy suggested there was an inequality involving a variable called "Full_window" being compared to the trigger threshold. We followed this down to another function in Report.cc where Full_window is used and compared fdiode_real between individuals, which was identical. We then tried printing V_Total_forconvlv, which was different between indviduals, yielding either all 0s or -nans in the individual with 0 effective volume.
Helped Ryan some more with working on the roulette algorithm in paperclip. We're going to take it step by step and start with getting a hang of the code with print statements and modifications to see how things work and adding in a function for the roulette algorithm that will simply select the individuals. Once we have that working, we can move onto the next step of properly implementing the roulette algorithm in the code in a manner similar to what we have in the bicone algorithm.
I made some small edits in the paper, mostly just pulling some citations from the proceeding but I also think I fixed the format of the references so that the numbers look nicer.
|
Alex P and I will continue looking in AraSim for the cause of the error. I'll also take some more time to contnue making edits to the paper. |
| Alex P |
Made a generation with an indiviudal we know got a zero fitness score and reduced the grid spacing to see if it would have any affect, it might stay zero as we've done this on a generation before and it lowered all the results so the outliers might not be the zeroes but instead the normal scores with the small antenna. Talked with Amy about looking for errors and started testing what gets passed into the myconvlv function and found that the fdiodes are equal between individuals but the V_Total_forconvlv array was different, and the indiviudal that had zero had a lot more "nans" than the one the had a real score. |
Continue investigating the source of where the changes in AraSim that allow for the zero fitness scores come from, marking off one variable at a time to find what makes the difference at each step |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Ran into more issues attempting to write in the genes for the roulette algorithm for paperclips. So, I'm taking a new approach to attempt to write a roulette function into the existing paperclips evolution to see If I can get it to work before I try to implement the paperclips genes into the roulette algorithm. |
Continue writing and attempting to run the roulette function inside of paperclips. |
|
|
|
67
|
Thu Jun 11 15:14:33 2020 |
Alex Patton | Daily Update 6/11/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
Helped Alex P look through AraSim some more. We wanted to keep trying to nail down whether or not the threshold was passed (the files said no before, but we wanted to try confirming by looking at the actual values). The files were big but from the values we saw none exceeded the threshold for the individuals with 0 effective volume. |
Amy has some more notes for the paper that I'll try to work on. I think we can try semi-running the loop to repeatedly pass in the same individuals and check the results with decreasing grid spacings from XF. |
| Alex P |
Worked with Alex M today to continue to comb through AraSim. Did in fact find the proper place where it checks the trigger and started by printing out the values it compares. When we printed all values instead of just the values that passed we got files that were 5-7 GB and didn't even finish, and weren't told much useful information since the failures were just 0 or -nan. Started by trying to work backwards and see where the value in the trigger comes from, but after looking through multiple files trying to trace back there ended up being a lot of possibilities of where it could have come from so we started looking from the top down and to look where the gainfile's values are used since we now know where the ending is it might be easier to find the path to the end from the start now. |
Continue to look through AraSim and trace the gainfile's path to affecting the trigger conditions to see what values would make it pass. Also continuing to investigate the grid spacing's affect on the gain file's values might be worthwhile after we know more about how those values matter. I do have a doctor's appointment tomorrow so will have to leave early and won't make it for group meeting. |
| Eliot |
Finished the plotting software to work and plot everything we want. Entire loop now ~should~ run in its entirety with asymmetry. Looking to do a couple of test runs over the next few days. Worked on CodeCademy git lesson. |
Continue work on git lesson and try to get a OSC interactive job. Make a few small edits that might be helpful from a user perspective. |
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
66
|
Wed Jun 10 15:47:43 2020 |
Alex Machtay | Daily Update 6/10/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M |
I wasn't available for most of the day, but I jumped onto the meeting to help Alex P continue looking for the problem with the effective volumes. Amy asked us to investigate the phase responses coming out of XF. I posted the plots in the drop box--see the text below this table for more details. |
I think we need to try a run of XF with variable grid spacings so that we can put in the same individuals with increasingly small grid spacings and get a clear view of how that affects the gain/phase vs frequency. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Continued working on the Roulette algorithm all the function definitions, constants and most of the main function are complete aside from parts that pertain to the genes of the paperclip antennas. |
If all goes well, I hope to find the gene parameters used in the paperclip tournament algorithm and then implement those into the work I did today and finish the roulette algorithm completely. If I complete that I plan on testing to make sure everything is working properly before merging onto the main development branch. |
For the phase vs frequency plots, see this link: https://www.dropbox.com/home/GP_Antennas/Updates/Phase%20vs%20Frequency%20plots .The plots show the average phase vs frequency for all 10 individuals in one generation of a run. The names of the plot files correspond to which run they come from.The runs which were examined were: XF_Data_Test, Grid_Space_Test, and Ryan_test_run3. They were chosen because they provide use with different antenna, grid spacing, and effective volume data.
The plot for XF_Data_Test is named p_vs_f_XFDT.png. The plot for Grid_Space_Test is named p_vs_f_GST.png. The plot for Ryan_test_run3 is named p_vs_f_Rtr3.png.
- XF_Data_Test a grid spacing of 0.1 cm. The antennas in generation 0 were large, but it evolved to make small antennas in generation 5, which is what the phase plot comes from. Individuals 7 and 10 had effective volumes of 0, while the remaining effective volumes were large.
- Grid_Space_Test used a grid spacing of 0.01 cm. The antennas in generation 0 were small, which is where the phase plot comes from. None of the individuals had 0 effective volume, but overall the effective volumes were lower than in the similarly sized antennas in generation 5 of XF_Data_Test.
- Ryan_test_run3 used a grid spacing of 0.1 cm. The antennas in generation 0, which is where the plot is from, were large and all had similar effective volumes to the actual bicone.
There are two things we noticed about these plots. First, they seem very noisy at low frequencies--they don't have much of a nice pattern here (actually, XF_Data_Test shows generally high phases at low frequencies that decrease with frequency up until around 200 MHz). The other detail we noticed is the periodic behavior of the average phase once it settles down beyond low frequencies. We don't know why this would occur. It's also worth noting that the phase diverge seemingly randomly at the last frequency--we assumed this was similar to how the phase is always wild at theta = 0 and theta = 360 in the .uan files. |
|
|
118
|
Thu Nov 19 16:42:06 2020 |
Alex Patton | Daily Update 11/19/20 | I hopped on with Alex M to catch him up with what has been done and changed within AraSim so far. I looked into using pointers to access arrays outside of their declared scope and it seemed reletively easy to set up. I set up a pointer corresponding to the arrays V_forfft, T_forfft, and vmmhz_filter and set them at the end of stage one in order to be used in stage two. Also made sure to delete the pointers at the end of stage 2 (Very Important). After this and finding the correct scope to put the deletes in, we complied and didn't get an error in Report.cc. Once it compiled I signed off to study for exam and Alex M will now take over and start to run tests to make sure that with the same seed this gives the same results as a base copy of AraSim. After this the next step would be to start implementing a way to save all the variables from stage one and reading them into stage 2. |
|
|
112
|
Fri Oct 9 17:37:57 2020 |
Alex Patton | Daily Update 10/9/20 |
| Name |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P, Ryan, and Ben |
Got all functions finished and worked on adding them to main. We set up their calls and commented out the unfunctionalized code with a comment explaining what it was and what date it was commented out. Had to define new variables for limits on our mutation function now that our mutation function is uniformly distributed within a range rather than using standard deviations. Also created some more global variables for maximum outer radius and minimum length in order to have them more accessible. |
Finish up implementing and defining variables so that it can run the functions properly. Hoping to have it ready to test on monday. |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ethan |
. |
|
|
|
|
117
|
Fri Oct 30 17:30:16 2020 |
Ethan Fahimi | Daily Update 10/30 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Attempted to make a new mutation function for the algorithm to try and address the concerns about hitting local maximums from Wednesday. Unfortunately, the idea was unsuccessful when I put it through testing. I would like some input from some of the experts before trying something else. Otherwise, the version I had earlier this week has still been very consistent about optimizing the runs outside of sometimes hitting a local max at about 90/100.
|
|
| Ben |
|
|
| Ethan |
Moved AREA onto my own directory with Alex's help. Began fixing issues with it (small win, no more permissions issues!). |
Continue fixing issues with AraSim on my user. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
|
Draft
|
Mon Nov 30 16:59:39 2020 |
Ethan Fahimi | Daily Update 10/30 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Attempted to make a new mutation function for the algorithm to try and address the concerns about hitting local maximums from Wednesday. Unfortunately, the idea was unsuccessful when I put it through testing. I would like some input from some of the experts before trying something else. Otherwise, the version I had earlier this week has still been very consistent about optimizing the runs outside of sometimes hitting a local max at about 90/100.
|
|
| Ben |
|
|
| Ethan |
Moved AREA onto my own directory with Alex's help. Began fixing issues with it (small win, no more permissions issues!). |
Continue fixing issues with AraSim on my user. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
|
116
|
Mon Oct 26 17:59:10 2020 |
Ethan Fahimi | Daily Update 10/26 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
I have the testing loop finished with plotted results now. The program was able to reach optimal results very quickly. It used 100% tournament selection with a cutoff on the outer radius. The algorithm was using an asymmetric algorithm and the ideal bicone it was being compared to was an arbitrarily picked one from an actual symmetric run individual we knew to stay within the outer radius. All individuals for each generation are plotted on this graph. And the fact that these bicones started as asymmetric shows that we can very easily find symmetric answers if they are indeed ideal. |
|
| Ben |
|
|
| Ethan |
Tried to fix permission issues with AREA, as well as learned about running the loop and listening to Jorge's thoughts on our project. |
Possibly move the AREA project onto the project space as it may solve permissions issues. Alex is looking into it. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
| Attachment 1: fitness.png
| 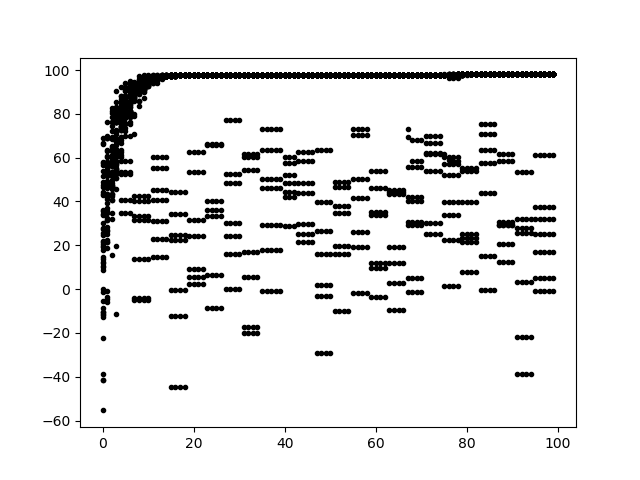 |
|
|
115
|
Mon Oct 19 17:02:12 2020 |
Ethan Fahimi | Daily Update 10/19 |
| Name |
Progress |
Plans |
| Alex M |
|
|
| Alex P |
|
|
| Ryan |
Alex P. and I put the outer radius constraint into the Asymmetric our version of the algorithm. I have also created the pseudo-fitness function to be able to do some optimization testing that bypasses the time-consuming parts of the run. All I need to do to finish the pseudo tests is to create a loop to run through the generations and plotting procedures. |
|
| Ben |
|
|
| Ethan |
Tried to fix permission issues, as well as work on running the loop (invalid id) |
Continue fixing AREA permission issues with Ben's help. |
| Parker |
|
|
| Elliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
|
|
|
Draft
|
Fri Oct 16 17:32:49 2020 |
Alex Patton | Daily Update 10/16/20 |
| |
Update |
Plans for Monday |
| Alex M |
|
|
| Alex P and Ryan |
Made new version of our modified GA that is asymetric. We didn't include the options to start with symetetry but just wanted to get it to a compilable state so that we can test how it would run in the loop to make sure it functions properly there. We also have a real run going with our previous symetrical GA edit. |
Watch this run over the weekend and keep it going and then eventually test how the asymetric version works |
| Eliot |
|
|
| Ben |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ethan |
Worked with Ben and Alex M on getting the AREA project working on slurm. It works now, but there are some permissions issues that we still need to fix. Simply put, Ben can run it perfectly right now. |
Find and address permissions issues so anyone can run a job. |
|
|
|
113
|
Mon Oct 12 17:34:29 2020 |
Alex Patton | Daily Update 10/12/20 |
| Name |
Update |
Plans for Week |
| Alex M |
|
|
| Alex P, Ryan, and Ben |
Finished up writing the updated GA and got it compilable and then got some run time segmentation faults but managed to get all of those fixed. But once we got it running we encountered an error where a lot of the generationDNA ended up being 0,0,0. We looked through our functions and confirmed this was only happening with indiviudals developed through crossover and not mutation or reproduction |
Get this problem fixed in crossover and continue to test and make sure this works as intended but so far it is able to generate genDNA files and just has that one bug. The bug is also every other individual so we think we might have messed up how crossover generates two individuals from parents so it should hopefully be an easy fix. |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ethan |
. |
|
|
|
|
55
|
Tue May 26 16:38:26 2020 |
Alex Machtay | Daily Update |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Continued working on fixing bugs with Alex P. I had a run from over the weekend which got through a few generations, but we're suspicious because the effective volumes are very high despite the antenna dimensions being very small. I made a python script that makes gain plots of the individuals and posted them in the dropbox. I'm not sure they tell us very much. We also got some individuals with effective volumes of 0 in the most recent generations. |
In the morning I'll be working with Julie, Alex P., Eliot, and Leo on fixing up some of the git stuff on Eliot's personal space. I also think I should start a run with small antennas but also a small grid size to see if this results in more realistic gain patterns that drive the effective volume down (or up for the ones which had 0 effective volume). |
|
Alex P.
|
Worked with Alex M to try to find bugs, looked through gain pattern plots but were unable to work out what the problem was. Also worked with Julie and others on getting git working so Eliot and Leo can start their work. |
Finish up getting Eliot and Leo set up and practice with git to make sure everything is in order and work on finding error. I want to save all the AraOut files so that we can go back and look at the events passed |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
58
|
Fri May 29 17:36:04 2020 |
Alex Machtay | Daily Update |
| Name |
Today's Update |
Plans for Monday |
| Alex M |
I wasn't available in the morning. I met with Amy and Alex P around 1 to talk about how to use the AraSim function for throwing the same neutrinos to search for bugs in the loop. We want to compare three types of antennas we've evolved to the actual ARA bicone: a small antenna with 0 effective volume, a small antenna with a normal effective volume, and a normal antenna with a normal effective volume.
I also spoke with Amy and Julie about the paper and worked on getting the paper in the right format, but I still can't get the citations to work.
|
I won't be available til late Monday morning, but I'll try to talk to Alex P about using the AraSim function and looking for bugs. Julie and I are supposed to meet at 1 with Ben to discuss him joining, and then I think we have a meeting at 1:30. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
76
|
Fri Jun 26 16:13:18 2020 |
Alex M | Daily Update |
| Name |
Today's Update |
Plans for Monday |
| Alex M |
Worked to run the loop with Alex P to test a smaller initial mean and standard deviation of the opening angle. Worked some more on the paper too--I fixed some citations issues (and found some good ones to add) but the references are still spilling into the margins and off the page. |
I'm going to look for where the 3D gain plots in XF for small antennas with different cone separations start to converge so that we can figure out what length we want to set as a cut off minimum to stop them from evolving to be so small. |
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Ryan |
|
|
| Evelyn |
(Finally) got Code Academy to work on my browser, started on the git tutotial |
Get as far into the git tutorial as I can |
| Ben |
|
|
| Ethan |
|
|
|
|
|
82
|
Mon Jul 6 16:49:21 2020 |
Alex M | Daily Update |
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M |
Fixed up a few more errors in the new roulette algorithm with the length cut. Started a run to test the loop.
Edited and added a few things to the paper.
|
My goal is to get a few generations done over night to look at in the morning. If it looks like things are working out, we're going to go ahead and start the process of merging (by adding our edits in to the database version fitst, then Eliot and Leo's asymmetric version, then merging on Wednesday hopefully). |
| Alex P |
Worked with Alex M to have our length test run and also helped ben with an issue with his code when submitted as a job |
Look at run in the morning and if it is good start merging and continuing to test the database feature on a variable amount of genes |
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
Continued refining the Roulette function in Paperclips. Now negative or zero fitness scores cannot be passed along and the roulette can be isolated from algorithm 1 in the code. |
For now, I am continuing to have the last two algorithms mutate for the rest of the individuals but am planning to write in the roulette to create new individuals that are mutated from the chosen. Once I complete this, I plan of coding in a way for user input to decide how many individuals are sent to each algorithm for selection and mutation. |
| Ben |
|
|
| Ethan |
|
|
|
|
|
Draft
|
Tue Aug 11 19:56:45 2020 |
Alex M | Daily Update |
| Name |
Update |
Plans for tomorrow |
| Alex M |
I worked on fixing the AraSim issue today. I think I have it resolved. Essentially, I made a second set of flags that the loop will look for and use the first set of flags to tell the loop which error files to look at to look for the phrase "segmentation fault." If it sees that phrase, it resubmits the job. Otherwise, it makes the second flag. Once all of the flags have been made, the loop can move on. I tested it by manually putting the phrase inside one of the error files to make sure it would catch it and resubmit the job.
I started a new run (partly to further test the fix, but primarily to get real data). It has the fixes we discussed last week at the GENETIS meeting (ex: mutations shouldn't repeat anymore).
|
I'm going to be out tomorrow and Thursday so someone needs to pick up my run. I'll make sure I open permissions tonight so that anyone can run it tomorrow. If you have questions you can slack me, but I probably won't be able to respond til super late.
If there are issues with AraSim jobs failing without resubmitting, see the instructions below.
|
| Alex P |
|
|
| Eliot |
|
|
| Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
| Ben |
|
|
| Ethan |
|
|
Instructions for persisting AraSim errors:
Even if we still have the problem where AraSim jobs fail and the fix doesn't catch them, you can still run the loop as long as you manually step back in the loop. Here's what you have to do:
1. Exit the loop (ctrl-c).
2. Clear the following directories in the RunName directory: AraSimFlags, AraSimConfirmed, (gen)_AraSim_Errors, (gen)_AraSim_Outputs.
3. Revert the save state (second number goes to 5). This let's you resubmit the AraSim job.
4. Run the loop again (make sure you're still in an interactive job). |
|
|
64
|
Mon Jun 8 15:16:16 2020 |
Alex Patton | Daily GENETIS Update 6/8/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Worked with Alex on finding the variables Amy suggested to us in AraSim. We found the Efield and max peak voltage variables and printed them, comparing three antennas: a small one with normal veffective (nonzero), a small on with 0 veffective, and the actual bicone. The electic field was the same for all antennas, but the peak voltages were all very low for the antenna with 0 veffective. Actually, after reinvestigating, it looks like the maximum peak voltages *were* the same between all the antennas.
I've been looking into using the automatic grid spacing in XF. I think I need to get a better understanding of XF, so I started playing with it to design things in the GUI. I want to be able to make a model using the xmacros that I can look at in the GUI so I can understand all of the functions being used. Since we've checked AraSim pretty extensively, we think that the issue is with XF. I think the grid spacing might be allowing small antennas to appear better than they are.
|
I think to investigate if the problem is in XF we should do a run of the same antenna with different grid spacing and look at the average gain plots. A preliminary test could be to try modeling a small antenna with a very large grid size--if that gives a very high gain, then it would suggest that the issue is with grid spacing being too large for small antennas.
I think I'll also keep playing with Karoo. I've been reviewing some ML material to refamiliarize myself and see if I can write a few simple programs. I got Karoo running on OSC last week but I don't really understand the outputs (using the practice dataset).
|
|
Alex P.
|
Worked through more with AraSim looking at Amy's suggestions she sent over the weekend. Was able to print the PeakV and the Electric Fields, the Electric Fields matched and were the same which is what should have happened and the PeakV varied depending on how well it detected the event. Also the trigger threshold is already printed so we looked at that too. |
We believe that AraSim is functioning properly so next we will look more into XF, and the grid spacing. ProGrid we think is the option from what we talked with Cade but we will continue to look further into it. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
Was added as a collaborator to the paperclip repository on git hub and created a new dev branch and directory. Afterward, I started writing the new roulette algorithm. All changes have been pushed to the githhub repository. |
Continue writing the roulette algorithm and start work on the github classes on Codecademy. |
|
|
|
63
|
Fri Jun 5 14:29:56 2020 |
Alex Patton | Daily GENETIS Update 6/5/20 |
| Name |
Today's Update |
Plans for Monday |
| Alex M. |
Helped Alex P try to figure out how to get data Amy was asking us to get for comparing antennas in AraSim. We've been stuck trying to figure out where all of the data is printed (there are a bunch of .cc files and quantities we aren't familiar with--for example, vmmhz in Report.cc sounds like the V/Mhz with the effective heights folded in, but we aren't sure).
I worked on some things in the paper that were mentioned in the minutes from last week's meeting, but I didn't see Amy's comments til the GENETIS meeting.
|
Monday morning Amy might stop by to talk to Alex and I about AraSim so we can try to find some of the info we haven't been able to print out. We'll also watch out for anything she posts in slack about it.
We'll present an outline of the paper at 12 on monday to the Ara group meeting to see if it needs to be an ARA paper or not.
I'm also going to keep looking for how to use the automatic grid spacing (and I'm planning on enlisting Eliot and Leo's help in being able to look at models in XF to make sure XF is designing antennas correctly).
|
|
Alex P.
|
Worked with Alex to find the volts/MHz after folding in the effective heights. While doing this we noticed a possible bug in AraSim. Currently our settings have a blank value for SIMULATION_MODE and a documentation we found says the default should be 0 but inside of AraSim the default is actually 1. Now Report.cc only uses volts/MHz if the SIMULATION MODE is set equal to 0 so this could've been an oversight when making the setup file if someone assumed leaving it blank would be zero. |
After checking whether the simulation mode is intended or an oversight, we want to check on the Volts/MHz and then also work on automatically setting the grid spacing and possibly work on implementing a penalty in the fitness score for antennas that get too small. |
| Eliot & Leo |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
61
|
Wed Jun 3 15:02:00 2020 |
Alex Patton | Daily GENETIS Update 6/3/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Talked with Alex about results from a run of small antennas where we decreased the grid spacing. The average gain plots look different, but I don't know yet if it's definitely resolved the problem because we're still getting a decent spread on the effective volumes. Helped Eliot and Leo with a git issue and talked to Evelyn and Ryan about paperclip. They were able to run and made some images, which is a good first step. I think they're going to start on making roulette an option since paperclip only uses tournament.
Julie gave me some more details about using Karoo with ANITA data.
|
I'll try installing Karoo and getting the old libraries that it needs. I should also try getting root to work on my personal space on OSC since I'll need that for the ANITA data (which I'm also going to need to find). I have a link to a dropbox with Oindree's stuff on blastfamy so I'll try looking there for hints. |
|
Alex P.
|
Ran average gain on smaller grid spacing for small antennas and did see pattern closer to larger antenna rather than the small antenna with larger grid spacing, also set up AraSim to print out HGain which should be the effective heights, which should run the next iteration of the loop. |
Compare HGain to find how th effective heights compare and make sure they do in fact differ between runs, also continue to see how grid spacing affects the small antenna and possibly run on a previous version which gave a zero vEff |
| Eliot & Leo |
Forgot to do updates this week so this is entire week. We have the loop running with two chromosomes up through Part C. ie we edited xmacros and other related parts to run simulation/XF with an asym bicone. Part D shouldn't need any changes, and Part E/F will need some.
|
Friday/through the weekend we will each do git on codecademy and next week we will implement changes in the fitness function and plottinf(Part E/F)
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
60
|
Tue Jun 2 13:42:21 2020 |
Alex Patton | Daily GENETIS Update 6/2/20 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Worked with Alex on trying to compare bicones to the ARA bicone. Met with Ben to give him some things to look over and told him to come to the meeting tomorrow at 1.
|
I might not be available all day tomorrow, but I should be good to come to the meeting at 1. Planning on doing a little bit of review of neural networks to get a feel of some of the stuff Julie is doing with Kai--I'm looking over notes and assignments from my ML class last fall. I also have a few things in the paper to fix up (namely citations, which I haven't been able to get to work). |
|
Alex P.
|
Ran analysis with bigger antennas and saw that they match the actual bicone more than the smaller ones in their overall average gain patterns, reduced grid spacing and started a run to see if it will change the outputs for the small antennas since we know that XF's generation of the UAN is the difference between gains and individuals. |
Observe results from the reduced grid spacing run and hopefully find some enlighting information to determine the next step. If not will try to find how to output electric field and compare that between our trials |
| Eliot & Leo |
|
|
| Evelyn |
We were able to find and run the old paperclip genetic algorithms, but we aren't completely sure how they work or what to do with the outputs. We also are not entirely sure which versions of the code to use, and were not able to find the code for a roulette algorithm. |
To find the roulette algorithm, and to learn exactly how the code that we already have works. |
| Ryan |
Found some executables for the paperclip algorithm and attempted to run them. Some worked, some did not, and we are currently still looking for any roulette algorithm and trying to figure out how to interpret the results. |
Continue searching for the Roulette algorithm and find out how to interpret the results. |
|
|
|
59
|
Mon Jun 1 15:21:03 2020 |
Alex Patton | Daily ELOG Update 6/1/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Spoke with Evelyn and Ryan about the paperclip antennas. They found the github repository with the files and a README so they're getting started on learning about the algorithm and testing out the scripts. Once they play with it and can get it running they'll start running it with different parameters for the selection methods (roulette vs tournament).
Also worked with Alex P. on figuring out the issue with the effective volumes coming out of AraSim. We have data from the eventReadIn function in AraSim for the small antennas and we don't see differences between the gains in these antennas and the actual bicone, but we still need to look at more of the quantities to compare that Amy told us about last week. We also came up with a new plot that might be interesting to look at. We think the plots look suspicious--I think it will be worthwhile to try rerunning with the same antennas but using a smaller grid spacing to see if the shape of the plots drastically changes.
|
Going to keep working with Alex P on resolving the small antenna effective volume issue. I'm also going to meet with Julie and Ben at 1 since Julie couldn't meet today. |
|
Alex P.
|
Continued to test with the Ara 0 vEff problem. Ran individuals with feed through events and individuals that gave 0 vEff had zero individuals pass still where others had consistent number pass. Used diff command and looked at AraOutput files and saw no difference except saying which events passed. Difference lies in .dat files passed into them which are generated from uan. These files are 130,000+ lines so we made a program to graph that average gain over the frequencies for the individuals and ran it with our small antennas. Interestingly, the ones that failed had higher average gains consistently than some runs that passed. Also ran this program with the Actual Bicone and the results were pretty different from the bicone and these small antenna. |
Next plan is to run our graphing code on a generation of previously large antennas generated to see if it makes any difference and maybe the problem is how these small antennas fitness scores get generated. |
| Eliot & Leo |
|
|
| Evelyn |
We were able to dig up the old paperclip code and make a new github repository with those files so we can work on it without breaking the originals. |
Figure out how to run the paperclip code from our own machines |
| Ryan |
Evelyn and I found the old paperclip repository on git, copied it over to our machines, and created a new repository to do our work on for the future. |
Create a dev branch and figure out how to run the algorithm. |
|
|
|
56
|
Wed May 27 15:00:28 2020 |
Alex Patton | Daily ELOG Update 5/27/2020 |
| Name |
Today's Update |
Plans for Tomorrow |
| Alex M. |
Met with Alex P. and Amy to think about how we can make sure the data coming from AraSim makes sense (specifically, to figure out why we're getting effective volumes of 0 on some antennas). Amy suggested we use a function in AraSim that let's us simulate identical events so that we can compare the same events between two different antennas. I asked Jorge in the #general chat to see if he could point us to where to look in AraSim for that function.
Met with Amy and Julie about people interested in joining the group and what upcoming project we have. We also talked about Karoo and Kai's thesis and how we can use machine learning to classify noise in the Anita data.
|
I won't be able to meet tomorrow or Friday morning, but I'll try to jump on in the afternoon on Friday and I should be able to make the GENETIS call. Also, Julie wanted to find a time where we could both meet with one of the new members if they have time this week. |
|
Alex P.
|
Worked in morning with Alex trying to find AraSim's 0 vEff problem, set it up to save AraOut files from now on in Run_Outputs in order to more easily look through. Continued with Git and successfully have working copies set up across multiple people and branches so we can all work without any conflicts. |
Continue to run and try to debug. Next idea is probably to set it up to pass exact events as specified so we know exactly what event is running and find error that way. |
| Eliot |
Today we finalized the GA. It now works with 2 chromosomes, where only length varies across chromosomes. In addition we got the tournament selection working (we don’t think it was working for even 1 chromosome prior). It is ready to begin being implemented tomorrow. We also met with Julie and others to discuss git and related topics.
|
Tomorrow we will implement the GA and begin editing Xmacros to 1) tell XF the new antenna type, etc and 2) have it read the new 2 chromosome data properly. This will likely take multiple days.
|
| Leo |
|
|
| Evelyn |
Learned more about git and github from Julie |
Find the old paperclip code and learn how to run and how it works! |
| Ryan |
Continued learning about Git and Github uses from Julie. Went back in the elog history to learn more about a previous "paperclip algorithm" for future uses. |
Learn more about the paperclip algorithm ad possibly help with more runs. |
|
|
|
54
|
Fri May 22 11:25:47 2020 |
Alex Machtay | Daily ELOG Update 5/22/20 |
| Name |
Update for today |
Plans for tomorrow |
| Alex M |
Worked with Alex P on fixing a new issue that appeared in AraSim. We were getting effective volumes of 0 in generation 4, despite getting nonzero fitness scores in previous generations. We made a fix that seems to give *some* non-zero effective volumes. The sequence of zero and non-zero fitness scores appears to correspond exactly with the length of the cones, but oddly it's preferring the shorter ones so I'm skeptical. |
I'm going to continue this run for another generation to see if generation 4 was a fluke, but because the change we made was substantial, we need to perform another run since previous generations may have been affected by this bug. |
| Alex P |
Worked a little bit to fix error with graphs submitting, worked with Alex M on fixing some problems and submitted long job of the ARA actual bicone so that we can get a worthy vEff from that change the graphing software to include it automatically rather than generating a new one each run. Started with just 100,000 neutrinos but hope to do a million but wanted to do 100,000 to see how long it takes before running something that large. Found XFintoARA error with Alex too that could've caused new 0 vEff error. |
Over the weekend check on the 100,000 neutrino run and possibly run a 1,000,000 run, but also continue to run our full runs over the weekend, possible focusing on getting our 9 generation database run up to 13 or more gens. |
| Leo |
Finished work on the 2 chromosome method. We fixed the issues with the roulette algorithm so that we get a second generation that we expect. |
Next week, Eliot and I hope to start looking, at and working with XF to see where we will need to make adjustments. |
| Eliot |
|
|
| Evelyn |
|
|
| Ryan |
|
|
|
|
|
239
|
Fri Sep 1 09:16:30 2023 |
Alex M | Curved Run With Realized Gain | We are beginning a new run with several improvements to the ARA VPol loop to try to evolve antennas that are optimally matched just by their geometry. To do this, we are evolving with the RealizedGain instead of just Gain in the Xmacros (thus taking into account impedance mismatch). We also have the speed up that parallelizes the AraSim jobs on each run. Attached is the run_details.txt file, but the GA parameters are subject to change. Here is what they are to begin:
We have a population size of 50 individuals per generation.
Selection operators (NUMBER selected by each):
- Roulette: 10
- Tournamente: 10
- Rank: 30
Genetic Operators (NUMBER generated by each):
- Reproduction: 6
- Crossover: 36
- Immigration: 8
- Mutation: 2
|
| Attachment 1: run_details_curved_realized.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_08_30_realized_curved' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=100 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=5 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=1500 ## Number of Neutrinos Thrown in AraSim
exp=18 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=4 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
RADIUS=0 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=0 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=0 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=0 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=1 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=6 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=2 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=10 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=10 ## Percent of individuals selected through tournament (divided by 10)
RANK=30 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
ParallelAra=1 ## Sets whether AraSim is being run on multiple threads or not
|
|
|
Draft
|
Mon Mar 6 00:07:32 2023 |
Dylan Wells | Current Status of PUEO and To Do List for Hackathon | I went through the current PUEO Loop and documented everything that still needs to be accomplished before we can run.
Everything is compiled in this Google Doc which I will paste the current version of below.
Current Loop Overview:
Part A:
Runs GA, need to implement most recent GA (easy)
Part B:
B1:
Enters PUEO freq into simulation_PEC.xmacro then cats in PUEO simulationPECmacroskeletons.
Will need to update simulationPECmacroskeletons with XF script to simulate the Hpol and Vpol sides of the antenna separately.
The bash portion should be working.
B2:
Currently almost the same as the ARA version. Creates the output.xmacro and moves the uan files. Will most likely need to change to get files in PUEO/icemc format, ready for input into the conversion script.
Part C:
Runs conversion script XFintoPUEO.py
The Bash portion is complete. We will need simple modifications to XFintoPUEO.py depending on XF outputs.
Part D:
D1:
Changes setup file, runs IceMCCall_Array.sh batch job
Need make this batch job to change IceMC / PUEO to read in the correct files
(vv_0_{gen}_{i}, hh_0_{gen}_{i}) for the antenna i in generation gen.
I’ve commented where this should go. The rest of the job should be complete
The debug mode for D1 might not work, but it shouldn’t be necessary to run.
So, the bash portion should be able to run.
D2:
Currently confirms runs by looking at the number of files in the AraSimConfirmed directory and looks for specific AraSim errors.
This line will need to be changed to count the output files for icemc/pueo.
Change the error detection to just resubmit the job or something similar as we aren’t yet familiar with icemc errors during runtime.
Part E:
Runs the PUEO fitness function. Unfortunately, icemc doesn’t output error bars, so we will have to remake any plots reliant on that. Veffective is also the same as fitness score.
Currently it should be working with the basic needed plots for gain patterns and veffective scores. But, we should add some more cool, automatic plots.
Part F:
The plotting software seems to be specific to ARA output, needs PUEO versions.
ASYM LOOP:
Should be working with PUEO as a switch
IMPORTANT ISSUES:
Find out how to simulate polarization in XF:
Modify the current XF model, can be found in PAS1960/dylanwells1629/testproject.xf
The current model has all 4 sides of the antenna connected. However, the actual antenna is basically 2 antennas, one hpol and one vpol, electrically disconnected from each other. So, adjacent parts shouldn’t be electrically connected like they currently are in the model. We might have to simulate the two models independently. The goal for this portion is to figure out how to split up the antenna in the xmacro, and how to either simulate hpol and vpol separately or how to get xf to output hpol and vpol gain patterns.
XF Cross - Polarization, constraints --Alex
Make IceMC / PUEO read in out input gain patterns
Will need to modify Batch_Jobs/IceMCCallArray.sh to change icemc to read in the current gain files before running. You can find the conversion script in Antenna_Performance_Metric/XFintoPUEO.py and the batch job will be run in Loop_Parts/Part_D
We need to change icemc / pueo to read in the correct gain files before running it.
This will involve changing the anitaBuildTool/components/icemc/anita.cc ReadGains function starting on line 1413 to read in vv_0_{gen}_{num}, hh_0_{gen}_{num} , etc. (Do this with PueoSim) -- Dylan
I’m not sure if you will need to recompile these functions before running the icemc executable again, so find that out too. (enter pueoBuolder/ just run make) --Dylan
Make PUEO plotting software
All of the current plotting software is found within Part_E and Part_F. Sadly, icemc does not output errors for the veffectives (at least not that I could find, maybe you can find it), so much of the Ara plotting software will not work with PUEO outputs. The fitness scores csv will be the same format as ARA’s, and veffectives will be the same as fitness scores for PUEO. Figure out how to change the existing ARA python plotting scripts or make new ones. --Bryan
Find out how to run PEUOsim and document outputs
Currently the loop is working with icemc, but we want to use PUEOsim for the future. So, document installing it, running it, and the outputs. (inputs are the same as icemc) -- Dylan |
|
|
130
|
Mon Mar 15 13:05:22 2021 |
Alex M | Current Run | We are currently running the loop using the improved parameters. The parameters are as follows:
50 individuals
80/20 Roulette/Tournament
6% Reproduction, 72% Crossover, 22% Mutation
Also note that individual 5 of generation 1 had its fitness score altered because one of its AraSim jobs didn't finish in time (and we would have had to wait 5 hours for it to rerun). To remedy this, I replaced the effective volume for the job with the average of the effective volumes from the other jobs for individual 5 (essentially meaning that it ran with 270k neutrinos instead of 300k). There is a .txt file in the directory titled Run_Notes.txt containing this message (and which will contain additional messages on similar things should they arise).
Run directory: /fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/Improved_Parameter_Run_2021_03_09 |
|
|
183
|
Mon Oct 24 16:07:22 2022 |
Dylan Wells | Creating the matching circuit design | The first part of designing the matching crictuit was choosing a frequency range to match over. We chose 100 MHz to 1000 MHz.
The geometric mean of this range in 316.227 MHz, which is the ideal frequeny to match to.
Then, we need the impedance of the antenna we are matching to.
The data for the best antenna is found in
/fs/project/PAS0654/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/AraSim_Polarity_Fix_2021_03_19/AraSim_Polarity_Fix_2021_03_19.xf/Simulations/001108/Run0001/output/SteadyStateOutput
The files begin at 83.33 MHz and increment by 16.66 MHz, so file 14 (316.57 MHz) is the best choice for matching.
This antenna has Impedance Z = 229.839 -i151.515 ohms
From here, we need to decide a design architecture for the matching circuit. Since the real component of the impedance is greater than the source's (50 ohms, the standard for electrical sources), a natural design is a lowpass downward circuit.
Next, we decide on a number of L networks to cascade in the circuit design, or how many 'rungs' in the ladder.
The purpose of these rungs is to artificially lower the ratio between the source impedance and the load impedance, as the closer these values, the broader the range of frequencies we can match.
The formula to calculate this is
N = ln(Rl/Rs)/ln(1+Q^2)
where
Q = ((Rl/Rs)-1)^½
Rs = real source impedance
Rl = real load impedance
For the given values, N is 14.
From here, we have all the information to find the capacitances and impedances for each L network in the ladder circuit.
The proccess of calciulating these values starts with finding the desired ratio between the impedances.
ratio = Q^2+1
Which we can use to create a list of 13 impedances between the source and load (corresponding to 14 rungs), each Q^2+1 times larger than the last. Then we can use the formula for a single lowpass downward match between the impedances of each rung.
All of this process is described in /users/PAS1960/dylanwells1629/improved_match_maker.py
The key functions PCSL, calcN, and broadbandMatchLP
|
|
|
187
|
Mon Dec 5 17:46:40 2022 |
Dylan Wells | Constraints on PUEO evolved antennas | Variables of Evolved Antennas
Wall variables:
S -- length of bottom of the walls
m -- slope of the walls (currently set to 1)
H -- height of walls
x0, y0, z0 -- 3D coordinates of the bottom of the ridge
xf, yf, zf -- 3D coordinates of the top of the ridge
Beta -- curvature of the ridge
Antenna Walls
S is half the side length of the bottom wall
m is the slope of the outer wall
H is the max height of the outer wall
Current Constraints:
S < 50cm
H < 50cm
m = 1
Antenna Ridges
x_0, y_0, z_0 are the innitial points of the inner most part of the ridge
x_f, y_f, z_f are the final points of the inner most part of the ridge
tau is the parametric time range beta is the slope of the curve of the ridge
Current Constraints:
tau = 0.26
x_f = S
0 < x_0 <= x_f
0 < y_0 <= x_0
0 < y_f < z_f
z_0 = 0
0 < z_f <= H
(4/30) * z_f < beta < 7 * z_f (for z_f in meters, designs will 'compile' in xfdtd above this upper limit, but the curve is functionally a line for all values greater) |
|
|
181
|
Thu Aug 18 13:03:47 2022 |
Dylan Wells | Comparing inputs and outputs between AraSim and IceMC | Comparing Inputs:
Frequency Lists:
Ara - 83.33MHz - 1066.70 MHz, step = 13.33MHz
IceMC - 200MHz - 1500MHz, step = 10MHz
Number of files read in:
Ara - 1
IceMC - 8
Formating of files:
Ara - Theta, Phi, Gain (dB, thetra), Gain (theta), Phase (theta)
IceMc - Frequency, Gain (dB) (Different files cover different thetas and phis)
File Type:
Ara - .txt
IceMC - no suffix (file with 2 columns of text)
Other notes for IceMC inputs:
IceMC reads in 8 different files for gain.
vv_0 hh_0 vh_0 hv_0 vv_el vv_az hh_el hh_az
Found in ../anitaBuiltTool/components/icemc/data
vv_0 = gains for vertical polarization
hh_0 = gains for horizontal polarization
vh_0 = gains for v → h cross polarization
hv_0 = gains for h → v cross polarization
vv_el = v polarization, e angle
vv_az = v polarization, a angle
hh_el = h polarization, e angle
hh_az = h polarization a angle
for e angle and a angle in
0. 0
-
5
-
10
-
20
-
30
-
45
-
90
(iterates 1 to 6)
Comparing Outputs:
IceMC - veff is in second column in veff+runName+.txt file (in the outputDirectory directory)
Ara - veff is at the bottom of the AraOut.txt file |
|
|
170
|
Tue Jun 28 13:27:13 2022 |
Dylan Wells | Changes needed for the matching circuit script |
-
Fix the functions for the SLPC, SCPL, and PLSC L networks (change the paramaters to match with the format of our data)
-
Write the PCSL function
-
Create a function to find the number of L networks necessary (N) given a source and load resistance as well as a frequency range.
-
Write a function to broadband match two impedances given a source, load, central frequency, and N. (return a list of capacitances and inductances for the L networks)
|
|
|
243
|
Mon Jun 2 14:10:59 2025 |
Jacob Weiler | Building Status 06/02/2025 | We are almost to where we can start the physical building of the antenna!
I've attached all the information I currently have regarding the building project. Some of it is messy work notes and some is well-structured.
I’ve attached the following files for the GENETIS building project:
- Building Dump.txt
- My working notes that I used while trying to simulate the antenna in XFdtd (very messy)
- Building Dump of Useful Materials.txt
- List of materials that I found regarding the building project like slides, elogs, etc.
- Simulating Building Model.txt
- A writeup I made describing my process for simulating the antenna in XFdtd
- Done with change materials.zip
- Solidworks model of antenna
I also made a slide deck that contains the directory locations + has graphs HERE. |
| Attachment 1: Building_Dump_of_Useful_Materials.txt
|
Building Dump:
For Initial Building Run:
Generation 13, individual 84 seems to be result being used (this assumption is based on the fact that when trying to straighten the sides for building they used this individual)
/fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_12_29
Elog Links for first building runs:
- Run Details: https://radiorm.physics.ohio-state.edu/elog/GENETIS/188
- Run Results + Gain Patterns: https://radiorm.physics.ohio-state.edu/elog/GENETIS/189
- Matching Circuit PCB: https://radiorm.physics.ohio-state.edu/elog/GENETIS/193
- Matching Circuit Parts: https://radiorm.physics.ohio-state.edu/elog/GENETIS/191
- Matching Circuit Schematic: https://radiorm.physics.ohio-state.edu/elog/GENETIS/230
- Matching Circuit Initial Design: https://radiorm.physics.ohio-state.edu/elog/GENETIS/183
- PoR Plots 1: https://radiorm.physics.ohio-state.edu/elog/GENETIS/194
- PoR Plots 2: https://radiorm.physics.ohio-state.edu/elog/GENETIS/196
- Straightened Sides 1: https://radiorm.physics.ohio-state.edu/elog/GENETIS/229
- Straightened Sides 2: https://radiorm.physics.ohio-state.edu/elog/GENETIS/236
- Engineering Call: https://docs.google.com/presentation/d/1Lo_6mFTmPbkToTrEeOpvznSdbxyPZexPag1qijTeYyM/edit?usp=sharing
At some point for some reason, another run seems to have been created for building with the crazy sides run here:
/fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_09_05_realized_curved_run
Top 5 vEffective Scores:
Value: 5.09897, Generation: 41, Individual: 44 (Seems to be this one, modified)
Value: 5.07746, Generation: 37, Individual: 16
Value: 5.05508, Generation: 37, Individual: 5
Value: 5.04558, Generation: 38, Individual: 12
Value: 5.04026, Generation: 48, Individual: 5
GENETIS Useful Links:
- GENETIS Google Drive: https://drive.google.com/drive/folders/1iDamk46R2_oOLHtvsOg4jNy05mCiB7Sn?dmr=1&ec=wgc-drive-hero-goto
- Onboarding Materials: https://radiorm.physics.ohio-state.edu/elog/GENETIS/41
- Julie's Dissertation: https://radiorm.physics.ohio-state.edu/elog/Write-Ups/220404_161525/Julie_Rolla_Dissertation.pdf
- Julie's Candidacy: https://as-phy-radiorm.asc.ohio-state.edu/elog/Write-Ups/44
- ICRC Proceedings: https://arxiv.org/pdf/2112.00197
- Phys Rev D Paper: https://journals.aps.org/prd/abstract/10.1103/PhysRevD.108.102002
- ARA Loop GitHub: https://github.com/osu-particle-astrophysics/GENETIS-ARA
- PUEO Loop GitHub: https://github.com/osu-particle-astrophysics/GENETIS_PUEO
- Shared Code GitHub: https://github.com/osu-particle-astrophysics/Shared-Code
- AraSim GitHub: https://github.com/ara-software/AraSim/tree/master
- pueoSim GitHub: https://github.com/PUEOCollaboration/pueoSim
|
| Attachment 2: Simulating_Building_Model.txt
|
Simulating Building Model
Getting the model we want to build from Solidworks into XF ready for simulations took a bit of work. Here are the steps and things I did to get it to finally work with materials and everything enabled (minus conductors in the coax cable).
General Instructions to setup the antenna the same as I did. Saving after each of these steps.
Getting out of Solidworks:
To get out of Solidworks, I used .step file under the assumption that it would carry the material data over into XF (this assumption was based on what I had read online, though I was looking at the wrong places for that information as I found out later). With this assumption, we spent time getting the materials correct in Solidworks before exporting out into the .step file. I spent considerable time double checking the materials in Solidworks to make sure that everything was defined correctly with at least good enough approximations of the materials to get a simulation working.
Material definitions:
Shells: Plastic wrapped in Copper Foil, approximated by just having the whole shell as Copper
Screws Connecting horizontal halves: ABS (PEEK Plastic)
Supports Connecting vertical halves: ABS
Other screws: Non-magnetic stainless steel (Passivated 18-8 Stainless Steel)
Coax Cable:
Dielectric: Foam Polyethylene (FPE)
Inner Conductor: Solid Bare Copper Covered Aluminum
Outer Conductor: Aluminum Tape
Outer Braid: Tinned Copper
Jacket: Polyethylene
Everything else: Approximated as copper (not entirely sure if they are copper fully or if they are just wrapped with copper foil)
Importing into XFdtd:
After having the step file exported, I put the file onto OSC and opened a new XFdtd project. I then clicked the "Import -> Cad Models" to select my file and have it imported. I did not import in the material data as I found out it did not import in correctly to each part, so I ignored it and manually added the material definitions later.
I now have the model into XFdtd, but it’s rotated 90 degrees to be in the horizontal plane. This isn’t inherently bad, but I want my surrounding scripts to not have to be changed much so I rotate the model to have the wire pointing in the +z direction in XFdtd. Once I’ve done this, I right click on the Braid and Inner + Outer conductors in the coax cable and select something similar to "Do not include in meshing." This now makes sure that these are NOT in the simulation.
Then, I manually added material properties into XFdtd from definitions I looked up online for the electric + magnetic properties of:
Copper
Plastic (ABS)
Foam Polyethylene (dielectric)
Polyethylene
Aluminum
Stainless Steel
After creating these material definitions, I applied them to the appropriate parts.
Feed adjustments:
I want the feed to be in the same location as the coax cable for the best results, problem is that there are holes in the place where the coax cable would be split (which I disabled to prevent shorting!). So, I setup two copper pucks (not much thicker than the copper pieces that cover the tops by the feeds) to fill out the holes and make sure each half is connected to each corresponding side of the feed. After I place these in the correct location, I use the same 50-ohm feed setup script used in the GENETIS Vpol loop.
Now we have everything almost ready to simulate.
Simulation Setup:
There are various things needed to be done to setup the script, and while you can use the GUI, I’m not familiar enough with it so I just used the corresponding scripts in the GENETIS loop that would be needed before an XF simulation takes place. After I run this, we are now ready to simulate.
Running Simulation:
Again, not familiar with the GUI so I just used the GENETIS XFdtd job scripts and modified them for this purpose (which was just adjusting directories of outputs). Then I submitted the job and waited for it to complete (I believe it took around 8 min per simulation for this antenna).
Getting uan files:
I then opened the simulation and ran the same code used to output UANs as used in the GENETIS loop to output all 60 uan files at the frequencies we want.
Now you should have the files for the building model that was made in CAD!
Debugging Steps I took:
This took me a while over spring break, at least a lot longer than I thought it would.
I found out that the material data from .step file does not translate as I had expected into XFdtd so I had to manually input the material data as shown above
I found out that hiding a part in XF does NOT exclude it from simulation, you have to remove it from meshing or it still remains there
I did compare the geometries between the as-evolved antenna and this building model, there are differences but they are slight. Overall they are very similar
Removing the conductors for the coax cable is necessary as it will just short the two pieces (leading back to 2) which makes sense
Final:
After doing all this, I ended up getting what I deemed reasonable for the outputs for the building model after 28 runs in my 03_13_2025_manual.xf xf file on my user. Run28 is the run that I describe setting up above this text.
The material is not 1-1 with what will be built as I found it difficult to find exact electro-magnetic properties for all of these, so maybe the discrepancies in gain could be resolved through more rigorous definitions. It could actually technically make it worse, but maybe when this is physically built this will need to be done to get more accurate results to compare against.
Simulating both of these with higher statistics in AraSim resulted in the antennas actually performing worse than the base Vpol antenna, which stinks but it is both of the antennas not just one!
After (delayed) emails back and forth with Christian Miki from University of Hawaii, he found these same issues while he was looking at the model from CAD before I went through the XFdtd simulation steps.
Material Definitions in XF:
For critique, here are the material definitions I used in the XF simulation (using XF material definition windows). You should be able to look at them in the actual xf project I mentioned above in my user (full path in slide decks)
All setup with the following:
Type: Physical
Electric: Isotropic
Magnetic: Isotropic
Passivated 18-8 Stainless Steel:
Electric Tab
Type: Nondispersive
Entry Method: Normal
Good Conductor: Automatic
Conductivity: 1.1e+06 S/m
Relative Permittivity: 1
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1.03
PEEK Plastic
Electric Tab
Type: Nondispersive
Entry Method: Loss Tangent
Good Conductor: Automatic
Relative Permittivity: 3.3
Loss Tangent: 0.003
Evaluation Frequency: 1 MHz
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1
Foam Polyethylene
Electric Tab
Type: Nondispersive
Entry Method: Loss Tangent
Good Conductor: Automatic
Relative Permittivity: 1.6
Loss Tangent: 0.0004
Evaluation Frequency: 1 MHz
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1
Polyethylene
Electric Tab
Type: Nondispersive
Entry Method: Loss Tangent
Good Conductor: Automatic
Relative Permittivity: 2.25
Loss Tangent: 0.0004
Evaluation Frequency: 1 MHz
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1
ABS Plastic
Electric Tab
Type: Nondispersive
Entry Method: Loss Tangent
Good Conductor: Automatic
Relative Permittivity: 3.2
Loss Tangent: 0.005
Evaluation Frequency: 1 MHz
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1
Copper Foil
Electric Tab
Type: Nondispersive
Entry Method: Normal
Good Conductor: Automatic
Conductivity: 5.96e+07 S/m
Relative Permittivity: 1
Infinite Dielectric Strength: Yes
Magnetic Tab
Type: Nondispersive
Entry Method: Normal
Conductivity: 0
Relative Permeability: 1
|
| Attachment 3: Building_Dump.txt
|
Building Dump:
Debugging Issues with Antenna model simulation:
Graphs to get (compared to Curved_Sides Antenna Run):
- Gain Plots
- Look at frequencies where dips. Could be due to: dielectric loss, mismatched impedance or structural changes
- Impedance Over Frequency Plots
- Want impedance to be around 50 Ohm for resistive components and 0 for reactance at operational frequencies
- S11 Plots (Return Loss VS Frequency)
- Look for where the S11 dips to determine where the antenna is resonant
- Total Efficiency Vs Frequencies
- Drops at certain frequencies indicates problems!
- VSWR vs Frequency
- Lower VSWR means better matching
in 03_13_2025_manual.xf:
Run1 = wrong material defs (deleted)
Run2 = glitched it
Run3 = wrong material def again slightly changed tho
Run4 = wrong material, with conductor gone
Run5 = wrong material, with full wire gone
Run6 = right material, full wire gone
Run7 = right material, conductor gone
Run8 = right material, everything there feed shifted to side
Run9 = right material, feed in middle of conductor
Run10 = right material, wire gone feed offset reduced (putting closer to center). this failed because the top of the feed was disconnected
Run11 = right material, feed with correct max feed offset allowed, coax gone
Run12 = trying the same thing but with the coax gone with building the feed
Run13 = with coax cable back, feed shifted closer to middle (apparently forgot to save and it's just the same thing.. as run12)
Run14 = adding pads and putting feed in the middle of the antenna, leave dielectric and jacket turned on
Run16 = pads, feed in middle, removing dielectric and jacket (-300 thing again.. not sure why)
Run17 = same thing but ABS material changed and adjusted pucks a little
Run18 = same ABS material change but with only inner conductor removed (I am testing why I am getting -300..)
Run19 = removed supports, still with pucks + only inner conductor removed
Run20 = removing pucks, with conductor removed and new ABS material (no more -300 but very low again...)
run21 = removed pucks, conductors(PLURAL) with new ABS Material
run23 = back to just supports, offset feed new ABS Material
run24 = coax gone, og ABS material, with the offset feed closer to the middle
run25 = everything back to normal coax gone (something wrong)
run26 = trying to fix the issue I'm seeing (FIXED) you have to uncheck that materials are included in meshing :/
run27 = actually removing the outer and inner conductors (yields worse gains!)
run28 = moving to feed center w/ copper plates and with the jacket + dielectric
Seems like the wire in the middle should be plastic (or non-conducting)? based off document wangjie sent me
"If we 3D print the metal, Chi-Chih thought that we could keep them together through a plasic rod running
through the middle" (It's not!)
maybe not, named LMR600 in solidworks which have the following material properties:
https://www.awcwire.com/lmr-cable/lmr-75-ohm-cable/lmr-600-75
screws connecting halves needed to be plastic
all other screws needed to be non-magnetic stainless steel
everything else is copper (?)
Trying to change materials of the wire and supports (03_13_2025_building_sim_2.xf): still bad
Trying again with same materials and putting feed down center of coax cable(03_13_2025_building_sim_3.xf): everything is -300 dBi :(
removing the copper middle part (03_13_2025_building_sim_4.xf): still bad
manually adding materials into XFdtd (03_13_2025_manual.xf): still bad, but different bad actually numbers-wise worse
- Passivated 18-8 Stainless Steel
- PEEK Plastic
- Dielectric: Foam Polyethylene (FPE)
- Inner Conductor: Solid Bare Copper Covered Aluminum
- Outer Conductor: Aluminum Tape
- Outer Braid: Tinned Copper
- Jacket: Polyethylene
- ABS Plastic
- Copper foil
I believe the feed replaces the coax cable in the middle so I am removing the inner conductor and assuming that it will be the same as the feed.
Dimensions of Curved Antenna (model based off this): (in cm for relevant parts)
- r1 = 3.20675
- height1 = 39.3683
- a1 = -0.0123505
- b1 = 0.418171
- r2 = 3.6116
- height2 = 18.605
- a2 = -0.0233028
- b2 = 0.369081
- Total height = 60.9733
Dimensions of Model in XF: (ignoring a's and b's as that's harder to measure..) (again in cm) (rough measurements in XF)
- r1 = 3.7
- h1 = 33.7441
- r2 = 3.4
- h2 = 18.71
- total height = 55.45 (no cable) 60.6459 (including cable)
Reference run XF settings:
- Removed the wire in the middle that was connecting the two sides: no difference (need to redo with it actually deleted + having the top plates copper) (03_11_2025_building_sim_1.xf)
- Removed middle wire AGAIN (03_13_2025_building_sim_0.xf): no difference, same issue
- Removed Supports and simulated(03_12_2025_building_sim_0.xf): This seems to have fixed the issue I'm seeing, so either the supports or the wire are shorting the antenna (or both!)
- Removed Supports ONLY(03_13_2025_building_sim_1.xf): still happening, though less extreme
For Initial Building Run:
Generation 13, individual 84 seems to be result being used (this assumption is based on the fact that when trying to straighten the sides for building they used this individual)
/fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_12_29
Elog Links for first building runs:
- Run Details: https://radiorm.physics.ohio-state.edu/elog/GENETIS/188
- Run Results + Gain Patterns: https://radiorm.physics.ohio-state.edu/elog/GENETIS/189
- Matching Circuit PCB: https://radiorm.physics.ohio-state.edu/elog/GENETIS/193
- Matching Circuit Parts: https://radiorm.physics.ohio-state.edu/elog/GENETIS/191
- Matching Circuit Schematic: https://radiorm.physics.ohio-state.edu/elog/GENETIS/230
- Matching Circuit Initial Design: https://radiorm.physics.ohio-state.edu/elog/GENETIS/183
- PoR Plots 1: https://radiorm.physics.ohio-state.edu/elog/GENETIS/194
- PoR Plots 2: https://radiorm.physics.ohio-state.edu/elog/GENETIS/196
- Straightened Sides 1: https://radiorm.physics.ohio-state.edu/elog/GENETIS/229
- Straightened Sides 2: https://radiorm.physics.ohio-state.edu/elog/GENETIS/236
At some point, another run seems to have been created for building with the crazy sides run here with REALIZED GAIN:
/fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_09_05_realized_curved_run
- Run is using the same freq of interest as what we currently use!!
Top 5 vEffective Scores of Realized Gain run:
Value: 5.09897, Generation: 41, Individual: 44 (Seems to be this one, modified)
Value: 5.07746, Generation: 37, Individual: 16
Value: 5.05508, Generation: 37, Individual: 5
Value: 5.04558, Generation: 38, Individual: 12
Value: 5.04026, Generation: 48, Individual: 5
GENETIS Useful Links:
- GENETIS Google Drive: https://drive.google.com/drive/folders/1iDamk46R2_oOLHtvsOg4jNy05mCiB7Sn?dmr=1&ec=wgc-drive-hero-goto
- Onboarding Materials: https://radiorm.physics.ohio-state.edu/elog/GENETIS/41
- Julie's Dissertation: https://radiorm.physics.ohio-state.edu/elog/Write-Ups/220404_161525/Julie_Rolla_Dissertation.pdf
- Julie's Candidacy: https://as-phy-radiorm.asc.ohio-state.edu/elog/Write-Ups/44
- ICRC Proceedings: https://arxiv.org/pdf/2112.00197
- Phys Rev D Paper: https://journals.aps.org/prd/abstract/10.1103/PhysRevD.108.102002
- ARA Loop GitHub: https://github.com/osu-particle-astrophysics/GENETIS-ARA
- PUEO Loop GitHub: https://github.com/osu-particle-astrophysics/GENETIS_PUEO
- Shared Code GitHub: https://github.com/osu-particle-astrophysics/Shared-Code
- AraSim GitHub: https://github.com/ara-software/AraSim/tree/master
- pueoSim GitHub: https://github.com/PUEOCollaboration/pueoSim
|
| Attachment 4: done_with_change_materials.zip
|
|
|
194
|
Mon Feb 13 14:56:57 2023 |
Jack Tillman | Building - Physics Plots for 9_50, 13_84, 18_89, 19_96, 29_87 Antennae | Physics plots were created for 300K events from the higher statistic simulation results for the 9_50, 13_84, 18_89, 19_96, and 29_87 antennae.
The simulation was inaccurate because a discrepancy exists between the higher simulation physics plots and the physics plots created for the 300K event results currently in the GENETIS Loop. This can be seen in the attached pdf file.
The simulation may have been inaccurate due to incorrect gain files.
The higher statistic simulations must be rerun for the 9_50, 13_84, 18_89, 19_96, and 29_87 antennae.
|
| Attachment 1: Genetis_Physics_Plot_Comparison_Slides.pdf
|
|
|
230
|
Thu Jul 6 14:52:09 2023 |
Jack Tillman | Building - Matching Circuit Schematic, PCB, and components | Attached are images of the matching circuit schematic and PCB design. A parts list is also attached in .pdf and .csv format. The .csv format can be imported into Digikey if necessary.
Table of component values:
| Inductors |
Capacitors |
SMA Connectors |
| 22 nH |
7.5 pF |
50 Ω |
| 27 nH |
5.7 pF |
50 Ω |
|
| Attachment 1: Matching_Circuit_Schematic.png
| 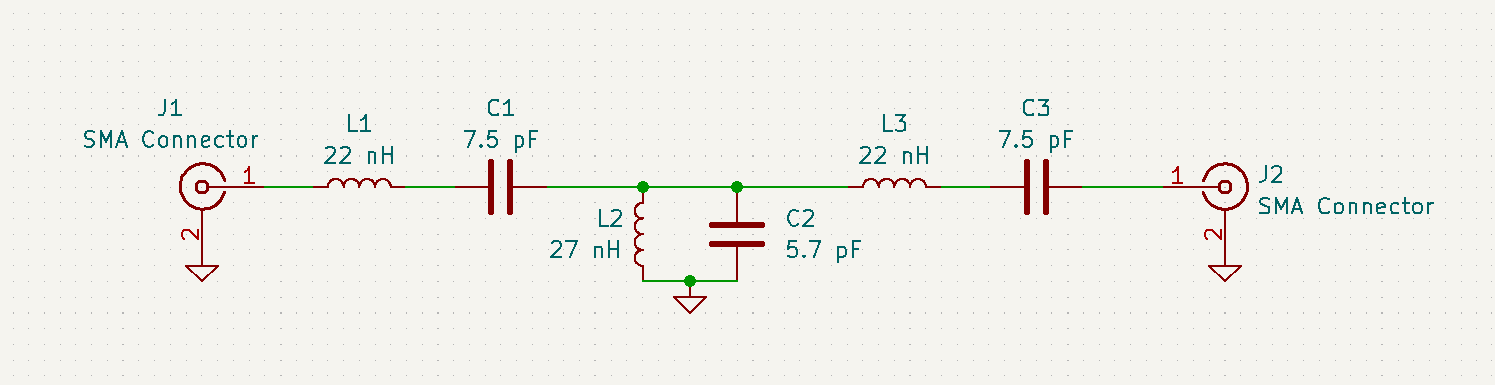 |
| Attachment 2: Matching_Circuit_PCB.png
| 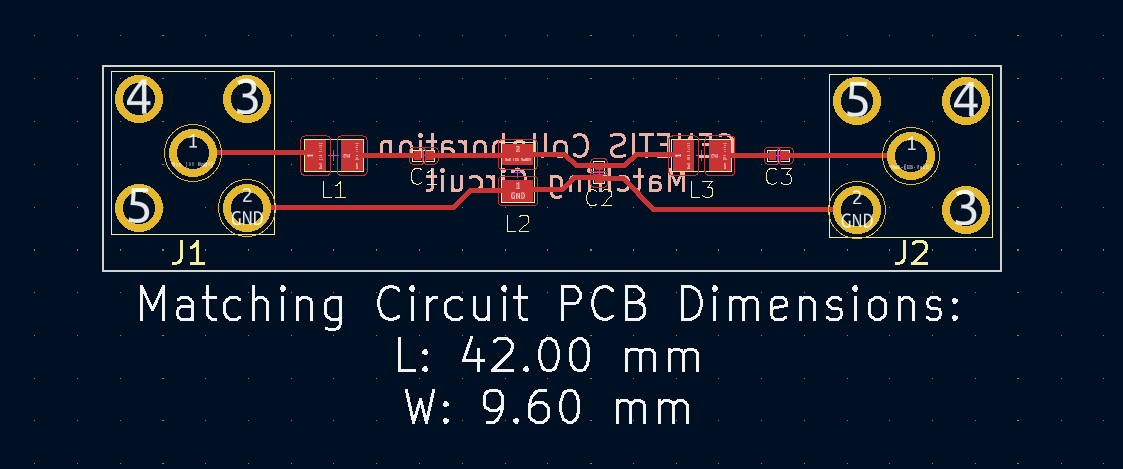 |
| Attachment 3: Matching_Circuit_DigiKey_Cart.pdf
|
| Attachment 4: Matching_Circuit_DigiKey_Cart.csv
|
Index,Quantity,Part Number,Manufacturer Part Number,Description,Available,Backorder,Unit Price,Extended Price USD
1,3,WM26450-ND,733910083,"SMA RA JACK, PCB",3,0,5.1,15.3
2,4,490-GJM0335C1E7R5BB01DCT-ND,GJM0335C1E7R5BB01D,CAP CER 7.5PF 25V C0G/NP0 0201,4,0,0.1,0.4
3,2,490-17198-1-ND,GJM0335C1H5R7CB01D,CAP CER 5.7PF 50V C0G/NP0 0201,2,0,0.09,0.18
4,2,490-16032-1-ND,LQW2BAN27NJ00L,FIXED IND 27NH 2A 70 MOHM SMD,2,0,0.42,0.84
5,4,490-16028-1-ND,LQW2BAN22NJ00L,FIXED IND 22NH 1.9A 70 MOHM SMD,4,0,0.42,1.68
|
|
|
193
|
Fri Feb 10 15:55:53 2023 |
Jack Tillman | Building - Matching Circuit PCB | I've completed laying out the PCB for the 14-rung matching circuit. Attached are png files of two PCB designs. One uses LPS5050 inductors while the other uses LPS6235 inductors. The dimensions shown are in millimeters. |
| Attachment 1: LPS5050_Dimensioned_PCB.png
| 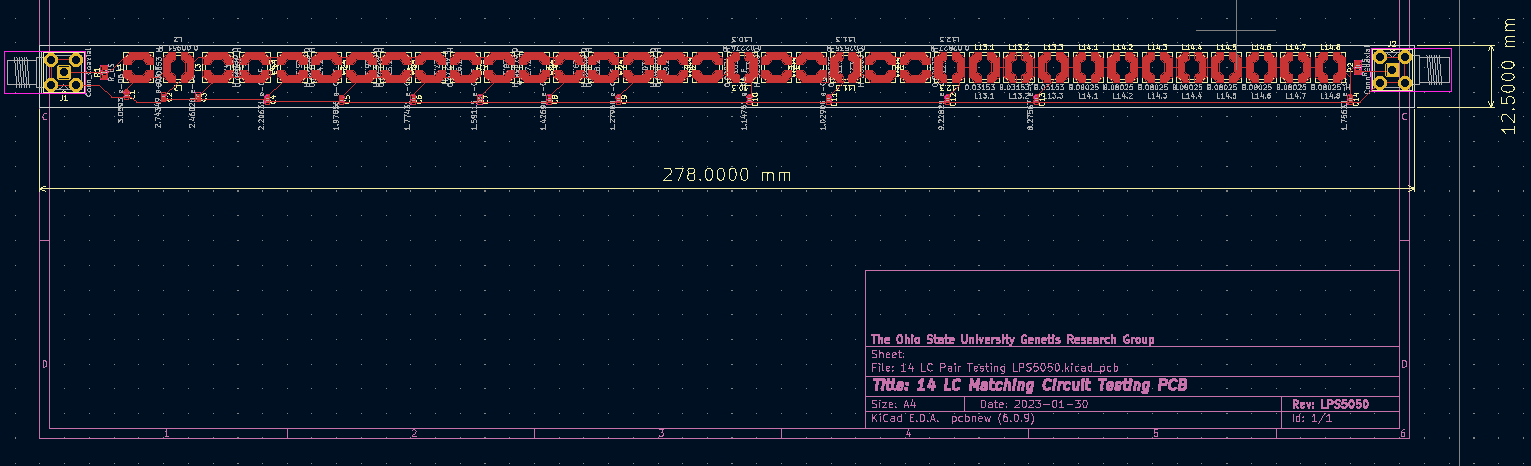 |
| Attachment 2: LPS6235_Dimensioned_PCB.png
| 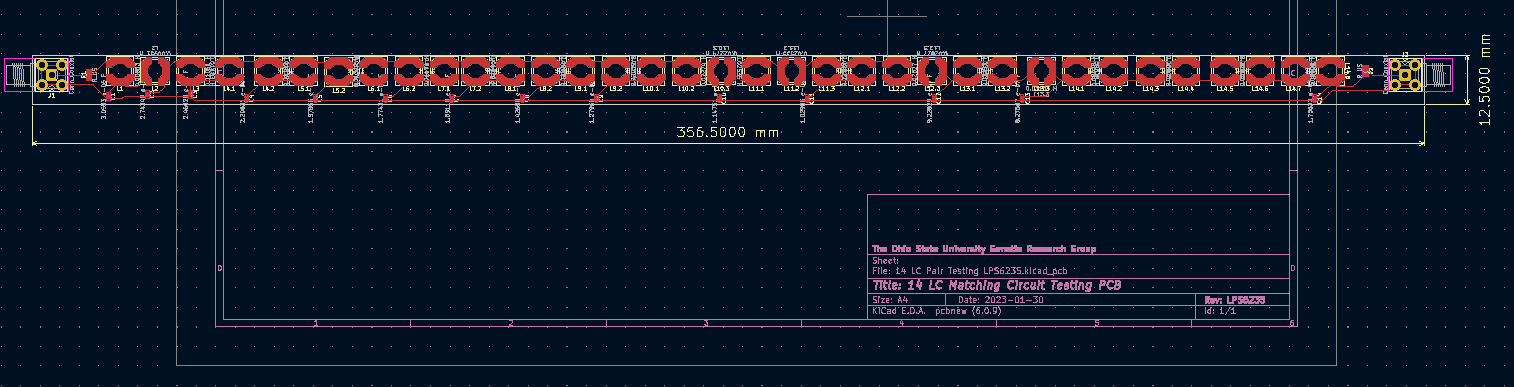 |
|
|
189
|
Tue Jan 31 11:30:54 2023 |
Audrey Zinn | Building | Attached are the XF files for the top 5 individuals from the 2022_12_29 crazy sides run. In order from best to 5th best:
1: Gen 29 Indiv 87
2: Gen 19 Indiv 96
3: Gen 13 Indiv 84
4: Gen 9 Indiv 50
5: Gen 18 Indiv 89
These can also be found in /fs/ess/PAS1960/BiconeEvolutionOSC/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2022_12_29/2022_12_29.xf in the corresponding directories. |
| Attachment 1: XF_model_13_84.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -64.90 0.00 -105.60
5 0 -19.63 0.01 -105.58
10 0 -13.64 0.04 -105.51
15 0 -10.17 0.10 -105.38
20 0 -7.75 0.17 -105.21
25 0 -5.90 0.26 -104.96
30 0 -4.43 0.36 -104.65
35 0 -3.23 0.48 -104.25
40 0 -2.22 0.60 -103.75
45 0 -1.38 0.73 -103.11
50 0 -0.67 0.86 -102.31
55 0 -0.08 0.98 -101.26
60 0 0.42 1.10 -99.87
65 0 0.82 1.21 -97.94
70 0 1.14 1.30 -95.09
75 0 1.39 1.38 -90.53
80 0 1.56 1.43 -82.17
85 0 1.67 1.47 -63.52
90 0 1.70 1.48 -19.53
95 0 1.67 1.47 24.56
100 0 1.56 1.43 43.26
105 0 1.39 1.38 51.64
110 0 1.14 1.30 56.22
115 0 0.82 1.21 59.07
120 0 0.41 1.10 61.01
125 0 -0.08 0.98 62.41
130 0 -0.68 0.86 63.46
135 0 -1.39 0.73 64.28
140 0 -2.23 0.60 64.92
145 0 -3.23 0.48 65.43
150 0 -4.44 0.36 65.84
155 0 -5.91 0.26 66.17
160 0 -7.76 0.17 66.42
165 0 -10.19 0.10 66.62
170 0 -13.66 0.04 66.75
175 0 -19.66 0.01 66.84
180 0 -64.87 0.00 66.87
0 5 -64.33 0.00 -107.33
5 5 -19.63 0.01 -107.32
10 5 -13.64 0.04 -107.27
15 5 -10.17 0.10 -107.18
20 5 -7.75 0.17 -107.05
25 5 -5.90 0.26 -106.88
30 5 -4.43 0.36 -106.65
35 5 -3.23 0.48 -106.37
40 5 -2.22 0.60 -106.01
45 5 -1.38 0.73 -105.57
50 5 -0.67 0.86 -105.00
55 5 -0.08 0.98 -104.28
60 5 0.42 1.10 -103.31
65 5 0.82 1.21 -101.97
70 5 1.14 1.30 -99.99
75 5 1.39 1.38 -96.77
80 5 1.56 1.43 -90.60
85 5 1.67 1.47 -74.77
90 5 1.70 1.48 -18.14
95 5 1.67 1.47 36.69
100 5 1.56 1.43 51.96
105 5 1.39 1.38 57.99
110 5 1.14 1.30 61.17
115 5 0.82 1.21 63.13
120 5 0.41 1.10 64.46
125 5 -0.08 0.98 65.43
130 5 -0.68 0.86 66.16
135 5 -1.39 0.73 66.73
140 5 -2.23 0.60 67.19
145 5 -3.23 0.48 67.56
150 5 -4.44 0.36 67.85
155 5 -5.91 0.26 68.09
160 5 -7.76 0.17 68.27
165 5 -10.19 0.10 68.42
170 5 -13.66 0.04 68.52
175 5 -19.66 0.01 68.58
180 5 -64.30 0.00 68.61
0 10 -63.85 0.00 -109.51
5 10 -19.63 0.01 -109.51
10 10 -13.64 0.04 -109.49
15 10 -10.17 0.10 -109.45
20 10 -7.75 0.17 -109.38
25 10 -5.90 0.26 -109.30
30 10 -4.43 0.36 -109.19
35 10 -3.23 0.48 -109.06
40 10 -2.22 0.60 -108.89
45 10 -1.38 0.73 -108.69
50 10 -0.67 0.86 -108.44
55 10 -0.08 0.98 -108.13
60 10 0.42 1.10 -107.73
65 10 0.82 1.21 -107.19
70 10 1.14 1.30 -106.41
75 10 1.39 1.38 -105.17
80 10 1.56 1.43 -102.75
85 10 1.67 1.47 -95.68
90 10 1.70 1.48 -10.72
95 10 1.67 1.47 57.68
100 10 1.56 1.43 64.05
105 10 1.39 1.38 66.34
110 10 1.14 1.30 67.55
115 10 0.82 1.21 68.31
120 10 0.41 1.10 68.85
125 10 -0.08 0.98 69.26
130 10 -0.68 0.86 69.58
135 10 -1.39 0.73 69.84
140 10 -2.23 0.60 70.06
145 10 -3.23 0.48 70.24
150 10 -4.44 0.36 70.39
155 10 -5.91 0.26 70.51
160 10 -7.76 0.17 70.61
165 10 -10.19 0.10 70.69
170 10 -13.66 0.04 70.75
175 10 -19.66 0.01 70.78
180 10 -63.82 0.00 70.80
0 15 -63.45 0.00 -112.40
5 15 -19.63 0.01 -112.41
10 15 -13.64 0.04 -112.42
15 15 -10.17 0.10 -112.44
20 15 -7.75 0.17 -112.47
25 15 -5.90 0.26 -112.50
30 15 -4.43 0.36 -112.55
35 15 -3.23 0.48 -112.61
40 15 -2.22 0.60 -112.70
45 15 -1.38 0.73 -112.83
50 15 -0.67 0.86 -113.01
55 15 -0.08 0.98 -113.26
60 15 0.42 1.10 -113.63
65 15 0.82 1.21 -114.19
70 15 1.14 1.30 -115.09
75 15 1.39 1.38 -116.67
80 15 1.56 1.43 -119.98
85 15 1.67 1.47 -130.06
90 15 1.70 1.48 150.28
95 15 1.67 1.47 88.84
100 15 1.56 1.43 80.40
105 15 1.39 1.38 77.44
110 15 1.14 1.30 75.99
115 15 0.82 1.21 75.16
120 15 0.41 1.10 74.65
125 15 -0.08 0.98 74.32
130 15 -0.68 0.86 74.09
135 15 -1.39 0.73 73.95
140 15 -2.23 0.60 73.85
145 15 -3.23 0.48 73.78
150 15 -4.44 0.36 73.74
155 15 -5.91 0.26 73.71
160 15 -7.76 0.17 73.70
165 15 -10.19 0.10 73.69
170 15 -13.66 0.04 73.69
175 15 -19.66 0.01 73.70
180 15 -63.42 0.00 73.71
0 20 -63.14 0.00 -116.43
5 20 -19.63 0.01 -116.46
10 20 -13.64 0.04 -116.53
15 20 -10.17 0.10 -116.63
20 20 -7.75 0.17 -116.78
25 20 -5.90 0.26 -116.98
30 20 -4.43 0.36 -117.25
35 20 -3.23 0.48 -117.59
40 20 -2.22 0.60 -118.04
45 20 -1.38 0.73 -118.62
50 20 -0.67 0.86 -119.39
55 20 -0.08 0.98 -120.42
60 20 0.42 1.10 -121.84
65 20 0.82 1.21 -123.88
70 20 1.14 1.30 -126.97
75 20 1.39 1.38 -132.07
80 20 1.56 1.43 -141.54
85 20 1.67 1.47 -162.14
90 20 1.70 1.48 155.74
95 20 1.67 1.47 117.62
100 20 1.56 1.43 100.14
105 20 1.39 1.38 91.86
110 20 1.14 1.30 87.29
115 20 0.82 1.21 84.47
120 20 0.41 1.10 82.60
125 20 -0.08 0.98 81.29
130 20 -0.68 0.86 80.35
135 20 -1.39 0.73 79.64
140 20 -2.23 0.60 79.12
145 20 -3.23 0.48 78.72
150 20 -4.44 0.36 78.41
155 20 -5.91 0.26 78.18
160 20 -7.76 0.17 78.02
165 20 -10.19 0.10 77.90
170 20 -13.66 0.04 77.82
175 20 -19.66 0.01 77.78
180 20 -63.11 0.00 77.78
0 25 -62.89 0.00 -122.51
5 25 -19.63 0.01 -122.57
10 25 -13.64 0.04 -122.71
15 25 -10.17 0.10 -122.95
20 25 -7.75 0.17 -123.28
25 25 -5.90 0.26 -123.72
30 25 -4.43 0.36 -124.31
35 25 -3.23 0.48 -125.05
40 25 -2.22 0.60 -126.00
45 25 -1.38 0.73 -127.22
50 25 -0.67 0.86 -128.80
55 25 -0.08 0.98 -130.86
60 25 0.42 1.10 -133.62
65 25 0.82 1.21 -137.41
70 25 1.14 1.30 -142.79
75 25 1.39 1.38 -150.72
80 25 1.56 1.43 -162.72
85 25 1.67 1.47 179.55
90 25 1.70 1.48 157.02
95 25 1.67 1.47 135.52
100 25 1.56 1.43 119.63
105 25 1.39 1.38 109.06
110 25 1.14 1.30 102.05
115 25 0.82 1.21 97.25
120 25 0.41 1.10 93.83
125 25 -0.08 0.98 91.34
130 25 -0.68 0.86 89.46
135 25 -1.39 0.73 88.03
140 25 -2.23 0.60 86.93
145 25 -3.23 0.48 86.07
150 25 -4.44 0.36 85.40
155 25 -5.91 0.26 84.89
160 25 -7.76 0.17 84.50
165 25 -10.19 0.10 84.22
170 25 -13.66 0.04 84.04
175 25 -19.66 0.01 83.94
180 25 -62.87 0.00 83.92
0 30 -62.72 0.00 -132.56
5 30 -19.63 0.01 -132.68
10 30 -13.64 0.04 -132.94
15 30 -10.17 0.10 -133.37
20 30 -7.75 0.17 -133.97
25 30 -5.90 0.26 -134.77
30 30 -4.43 0.36 -135.79
35 30 -3.23 0.48 -137.09
40 30 -2.22 0.60 -138.71
45 30 -1.38 0.73 -140.72
50 30 -0.67 0.86 -143.24
55 30 -0.08 0.98 -146.39
60 30 0.42 1.10 -150.34
65 30 0.82 1.21 -155.35
70 30 1.14 1.30 -161.66
75 30 1.39 1.38 -169.57
80 30 1.56 1.43 -179.20
85 30 1.67 1.47 169.63
90 30 1.70 1.48 157.66
95 30 1.67 1.47 145.96
100 30 1.56 1.43 135.50
105 30 1.39 1.38 126.73
110 30 1.14 1.30 119.66
115 30 0.82 1.21 114.04
120 30 0.41 1.10 109.61
125 30 -0.08 0.98 106.09
130 30 -0.68 0.86 103.30
135 30 -1.39 0.73 101.07
140 30 -2.23 0.60 99.29
145 30 -3.23 0.48 97.86
150 30 -4.44 0.36 96.72
155 30 -5.91 0.26 95.83
160 30 -7.76 0.17 95.15
165 30 -10.19 0.10 94.66
170 30 -13.66 0.04 94.33
175 30 -19.66 0.01 94.15
180 30 -62.70 0.00 94.13
0 35 -62.62 0.00 -150.86
5 35 -19.63 0.01 -151.07
10 35 -13.64 0.04 -151.49
15 35 -10.17 0.10 -152.14
20 35 -7.75 0.17 -153.03
25 35 -5.90 0.26 -154.18
30 35 -4.43 0.36 -155.60
35 35 -3.23 0.48 -157.32
40 35 -2.22 0.60 -159.38
45 35 -1.38 0.73 -161.80
50 35 -0.67 0.86 -164.62
55 35 -0.08 0.98 -167.87
60 35 0.42 1.10 -171.57
65 35 0.82 1.21 -175.73
70 35 1.14 1.30 179.66
75 35 1.39 1.38 174.64
80 35 1.56 1.43 169.28
85 35 1.67 1.47 163.72
90 35 1.70 1.48 158.08
95 35 1.67 1.47 152.52
100 35 1.56 1.43 147.18
105 35 1.39 1.38 142.18
110 35 1.14 1.30 137.59
115 35 0.82 1.21 133.45
120 35 0.41 1.10 129.78
125 35 -0.08 0.98 126.55
130 35 -0.68 0.86 123.75
135 35 -1.39 0.73 121.34
140 35 -2.23 0.60 119.29
145 35 -3.23 0.48 117.57
150 35 -4.44 0.36 116.14
155 35 -5.91 0.26 114.99
160 35 -7.76 0.17 114.09
165 35 -10.19 0.10 113.43
170 35 -13.66 0.04 113.00
175 35 -19.66 0.01 112.78
180 35 -62.60 0.00 112.77
0 40 -62.58 0.00 177.48
... 159721 more lines ...
|
| Attachment 2: XF_model_9_50.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -65.66 0.00 68.97
5 0 -23.00 0.01 68.99
10 0 -17.02 0.02 69.05
15 0 -13.55 0.04 69.15
20 0 -11.13 0.08 69.30
25 0 -9.29 0.12 69.49
30 0 -7.82 0.17 69.75
35 0 -6.63 0.22 70.06
40 0 -5.63 0.27 70.43
45 0 -4.79 0.33 70.90
50 0 -4.09 0.39 71.47
55 0 -3.50 0.45 72.18
60 0 -3.01 0.50 73.09
65 0 -2.61 0.55 74.31
70 0 -2.29 0.59 76.06
75 0 -2.05 0.62 78.83
80 0 -1.88 0.65 84.06
85 0 -1.78 0.66 97.84
90 0 -1.74 0.67 159.48
95 0 -1.78 0.66 -139.66
100 0 -1.88 0.65 -126.04
105 0 -2.05 0.62 -120.85
110 0 -2.30 0.59 -118.08
115 0 -2.61 0.55 -116.34
120 0 -3.02 0.50 -115.12
125 0 -3.51 0.45 -114.20
130 0 -4.10 0.39 -113.48
135 0 -4.80 0.33 -112.91
140 0 -5.63 0.27 -112.44
145 0 -6.63 0.22 -112.05
150 0 -7.83 0.16 -111.73
155 0 -9.30 0.12 -111.47
160 0 -11.14 0.08 -111.26
165 0 -13.57 0.04 -111.10
170 0 -17.04 0.02 -110.98
175 0 -23.04 0.00 -110.91
180 0 -65.62 0.00 -110.88
0 5 -66.34 0.00 70.70
5 5 -23.00 0.01 70.72
10 5 -17.02 0.02 70.80
15 5 -13.55 0.04 70.93
20 5 -11.13 0.08 71.12
25 5 -9.29 0.12 71.39
30 5 -7.82 0.17 71.72
35 5 -6.63 0.22 72.14
40 5 -5.63 0.27 72.65
45 5 -4.79 0.33 73.29
50 5 -4.09 0.39 74.09
55 5 -3.50 0.45 75.11
60 5 -3.01 0.50 76.44
65 5 -2.61 0.55 78.24
70 5 -2.29 0.59 80.87
75 5 -2.05 0.62 85.05
80 5 -1.88 0.65 92.78
85 5 -1.78 0.66 110.94
90 5 -1.74 0.67 160.03
95 5 -1.78 0.66 -152.08
100 5 -1.88 0.65 -134.51
105 5 -2.05 0.62 -126.96
110 5 -2.30 0.59 -122.85
115 5 -2.61 0.55 -120.26
120 5 -3.02 0.50 -118.46
125 5 -3.51 0.45 -117.14
130 5 -4.10 0.39 -116.13
135 5 -4.80 0.33 -115.32
140 5 -5.63 0.27 -114.67
145 5 -6.63 0.22 -114.15
150 5 -7.83 0.16 -113.72
155 5 -9.30 0.12 -113.38
160 5 -11.14 0.08 -113.10
165 5 -13.57 0.04 -112.89
170 5 -17.04 0.02 -112.75
175 5 -23.04 0.00 -112.66
180 5 -66.29 0.00 -112.62
0 10 -67.14 0.00 72.17
5 10 -23.00 0.01 72.19
10 10 -17.02 0.02 72.28
15 10 -13.55 0.04 72.45
20 10 -11.13 0.08 72.68
25 10 -9.29 0.12 72.99
30 10 -7.82 0.17 73.40
35 10 -6.63 0.22 73.91
40 10 -5.63 0.27 74.54
45 10 -4.79 0.33 75.33
50 10 -4.09 0.39 76.32
55 10 -3.50 0.45 77.59
60 10 -3.01 0.50 79.26
65 10 -2.61 0.55 81.54
70 10 -2.29 0.59 84.86
75 10 -2.05 0.62 90.10
80 10 -1.88 0.65 99.46
85 10 -1.78 0.66 119.21
90 10 -1.74 0.67 160.25
95 10 -1.78 0.66 -159.82
100 10 -1.88 0.65 -140.91
105 10 -2.05 0.62 -131.87
110 10 -2.30 0.59 -126.77
115 10 -2.61 0.55 -123.52
120 10 -3.02 0.50 -121.27
125 10 -3.51 0.45 -119.61
130 10 -4.10 0.39 -118.35
135 10 -4.80 0.33 -117.36
140 10 -5.63 0.27 -116.57
145 10 -6.63 0.22 -115.93
150 10 -7.83 0.16 -115.41
155 10 -9.30 0.12 -114.99
160 10 -11.14 0.08 -114.67
165 10 -13.57 0.04 -114.42
170 10 -17.04 0.02 -114.25
175 10 -23.04 0.00 -114.14
180 10 -67.09 0.00 -114.10
0 15 -68.08 0.00 73.45
5 15 -23.00 0.01 73.48
10 15 -17.02 0.02 73.58
15 15 -13.55 0.04 73.77
20 15 -11.13 0.08 74.04
25 15 -9.29 0.12 74.40
30 15 -7.82 0.17 74.86
35 15 -6.63 0.22 75.45
40 15 -5.63 0.27 76.18
45 15 -4.79 0.33 77.10
50 15 -4.09 0.39 78.26
55 15 -3.50 0.45 79.74
60 15 -3.01 0.50 81.70
65 15 -2.61 0.55 84.37
70 15 -2.29 0.59 88.24
75 15 -2.05 0.62 94.27
80 15 -1.88 0.65 104.69
85 15 -1.78 0.66 124.77
90 15 -1.74 0.67 160.35
95 15 -1.78 0.66 -165.00
100 15 -1.88 0.65 -145.87
105 15 -2.05 0.62 -135.89
110 15 -2.30 0.59 -130.06
115 15 -2.61 0.55 -126.29
120 15 -3.02 0.50 -123.67
125 15 -3.51 0.45 -121.74
130 15 -4.10 0.39 -120.28
135 15 -4.80 0.33 -119.13
140 15 -5.63 0.27 -118.21
145 15 -6.63 0.22 -117.47
150 15 -7.83 0.16 -116.88
155 15 -9.30 0.12 -116.40
160 15 -11.14 0.08 -116.03
165 15 -13.57 0.04 -115.75
170 15 -17.04 0.02 -115.55
175 15 -23.04 0.00 -115.43
180 15 -68.02 0.00 -115.39
0 20 -69.19 0.00 74.59
5 20 -23.00 0.01 74.62
10 20 -17.02 0.02 74.74
15 20 -13.55 0.04 74.95
20 20 -11.13 0.08 75.25
25 20 -9.29 0.12 75.65
30 20 -7.82 0.17 76.17
35 20 -6.63 0.22 76.82
40 20 -5.63 0.27 77.64
45 20 -4.79 0.33 78.67
50 20 -4.09 0.39 79.97
55 20 -3.50 0.45 81.64
60 20 -3.01 0.50 83.84
65 20 -2.61 0.55 86.85
70 20 -2.29 0.59 91.16
75 20 -2.05 0.62 97.79
80 20 -1.88 0.65 108.88
85 20 -1.78 0.66 128.73
90 20 -1.74 0.67 160.41
95 20 -1.78 0.66 -168.71
100 20 -1.88 0.65 -149.83
105 20 -2.05 0.62 -139.26
110 20 -2.30 0.59 -132.89
115 20 -2.61 0.55 -128.71
120 20 -3.02 0.50 -125.78
125 20 -3.51 0.45 -123.62
130 20 -4.10 0.39 -121.98
135 20 -4.80 0.33 -120.69
140 20 -5.63 0.27 -119.66
145 20 -6.63 0.22 -118.84
150 20 -7.83 0.16 -118.18
155 20 -9.30 0.12 -117.66
160 20 -11.14 0.08 -117.24
165 20 -13.57 0.04 -116.93
170 20 -17.04 0.02 -116.72
175 20 -23.04 0.00 -116.58
180 20 -69.13 0.00 -116.53
0 25 -70.51 0.00 75.63
5 25 -23.00 0.01 75.67
10 25 -17.02 0.02 75.80
15 25 -13.56 0.04 76.02
20 25 -11.13 0.08 76.35
25 25 -9.29 0.12 76.79
30 25 -7.82 0.17 77.35
35 25 -6.63 0.22 78.07
40 25 -5.63 0.27 78.97
45 25 -4.79 0.33 80.10
50 25 -4.09 0.39 81.52
55 25 -3.50 0.45 83.36
60 25 -3.01 0.50 85.77
65 25 -2.61 0.55 89.05
70 25 -2.29 0.59 93.74
75 25 -2.05 0.62 100.82
80 25 -1.88 0.65 112.32
85 25 -1.78 0.66 131.71
90 25 -1.74 0.67 160.43
95 25 -1.78 0.66 -171.53
100 25 -1.88 0.65 -153.09
105 25 -2.05 0.62 -142.16
110 25 -2.30 0.59 -135.37
115 25 -2.61 0.55 -130.85
120 25 -3.02 0.50 -127.66
125 25 -3.51 0.45 -125.31
130 25 -4.10 0.39 -123.51
135 25 -4.80 0.33 -122.10
140 25 -5.63 0.27 -120.98
145 25 -6.63 0.22 -120.09
150 25 -7.83 0.16 -119.37
155 25 -9.30 0.12 -118.79
160 25 -11.14 0.08 -118.35
165 25 -13.57 0.04 -118.01
170 25 -17.04 0.02 -117.77
175 25 -23.04 0.00 -117.63
180 25 -70.44 0.00 -117.58
0 30 -72.08 0.00 76.59
5 30 -23.00 0.01 76.63
10 30 -17.02 0.02 76.77
15 30 -13.56 0.04 77.02
20 30 -11.13 0.08 77.37
25 30 -9.29 0.12 77.84
30 30 -7.82 0.17 78.45
35 30 -6.63 0.22 79.23
40 30 -5.63 0.27 80.20
45 30 -4.79 0.33 81.41
50 30 -4.09 0.39 82.96
55 30 -3.50 0.45 84.93
60 30 -3.01 0.50 87.53
65 30 -2.61 0.55 91.06
70 30 -2.29 0.59 96.04
75 30 -2.05 0.62 103.48
80 30 -1.88 0.65 115.21
85 30 -1.78 0.66 134.04
90 30 -1.74 0.67 160.43
95 30 -1.78 0.66 -173.76
100 30 -1.88 0.65 -155.84
105 30 -2.05 0.62 -144.70
110 30 -2.30 0.59 -137.59
115 30 -2.61 0.55 -132.79
120 30 -3.02 0.50 -129.38
125 30 -3.51 0.45 -126.85
130 30 -4.10 0.39 -124.91
135 30 -4.80 0.33 -123.40
140 30 -5.63 0.27 -122.20
145 30 -6.63 0.22 -121.23
150 30 -7.83 0.16 -120.46
155 30 -9.30 0.12 -119.84
160 30 -11.14 0.08 -119.36
165 30 -13.57 0.04 -119.00
170 30 -17.04 0.02 -118.75
175 30 -23.04 0.00 -118.60
180 30 -72.01 0.00 -118.54
0 35 -73.95 0.00 77.50
5 35 -23.00 0.01 77.55
10 35 -17.02 0.02 77.70
15 35 -13.56 0.04 77.95
20 35 -11.13 0.08 78.33
25 35 -9.29 0.12 78.83
30 35 -7.82 0.17 79.49
35 35 -6.63 0.22 80.31
40 35 -5.63 0.27 81.35
45 35 -4.79 0.33 82.65
50 35 -4.09 0.39 84.29
55 35 -3.50 0.45 86.40
60 35 -3.01 0.50 89.17
65 35 -2.61 0.55 92.90
70 35 -2.29 0.59 98.15
75 35 -2.05 0.62 105.86
80 35 -1.88 0.65 117.70
85 35 -1.78 0.66 135.93
90 35 -1.74 0.67 160.42
95 35 -1.78 0.66 -175.60
100 35 -1.88 0.65 -158.22
105 35 -2.05 0.62 -146.97
110 35 -2.30 0.59 -139.62
115 35 -2.61 0.55 -134.58
120 35 -3.02 0.50 -130.97
125 35 -3.51 0.45 -128.29
130 35 -4.10 0.39 -126.23
135 35 -4.80 0.33 -124.62
140 35 -5.63 0.27 -123.33
145 35 -6.63 0.22 -122.31
150 35 -7.83 0.16 -121.49
155 35 -9.30 0.12 -120.83
160 35 -11.14 0.08 -120.32
165 35 -13.57 0.04 -119.94
170 35 -17.04 0.02 -119.67
175 35 -23.04 0.00 -119.51
180 35 -73.87 0.00 -119.46
0 40 -76.10 0.00 78.37
... 159721 more lines ...
|
| Attachment 3: XF_model_18_89.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -300.00 0.00 -25.63
5 0 -300.00 0.00 -25.69
10 0 -300.00 0.00 -25.72
15 0 -300.00 0.00 -25.73
20 0 -300.00 0.00 -25.71
25 0 -300.00 0.00 -25.66
30 0 -300.00 0.00 -25.58
35 0 -300.00 0.00 -25.48
40 0 -300.00 0.00 -25.35
45 0 -300.00 0.00 -25.20
50 0 -300.00 0.00 -25.02
55 0 -300.00 0.00 -24.82
60 0 -300.00 0.00 -24.59
65 0 -300.00 0.00 -24.33
70 0 -300.00 0.00 -24.06
75 0 -300.00 0.00 -23.76
80 0 -300.00 0.00 -23.44
85 0 -300.00 0.00 -23.11
90 0 -300.00 0.00 -22.76
95 0 -300.00 0.00 -22.39
100 0 -300.00 0.00 -22.02
105 0 -300.00 0.00 -21.63
110 0 -300.00 0.00 -21.24
115 0 -300.00 0.00 -20.85
120 0 -300.00 0.00 -20.46
125 0 -300.00 0.00 -20.07
130 0 -300.00 0.00 -19.70
135 0 -300.00 0.00 -19.34
140 0 -300.00 0.00 -18.99
145 0 -300.00 0.00 -18.67
150 0 -300.00 0.00 -18.37
155 0 -300.00 0.00 -18.09
160 0 -300.00 0.00 -17.85
165 0 -300.00 0.00 -17.64
170 0 -300.00 0.00 -17.47
175 0 -300.00 0.00 -17.33
180 0 -300.00 0.00 -17.23
0 5 -300.00 0.00 -57.01
5 5 -300.00 0.00 -56.84
10 5 -300.00 0.00 -56.42
15 5 -300.00 0.00 -55.73
20 5 -300.00 0.00 -54.77
25 5 -300.00 0.00 -53.53
30 5 -300.00 0.00 -52.01
35 5 -300.00 0.00 -50.20
40 5 -300.00 0.00 -48.07
45 5 -300.00 0.00 -45.62
50 5 -300.00 0.00 -42.84
55 5 -300.00 0.00 -39.72
60 5 -300.00 0.00 -36.29
65 5 -300.00 0.00 -32.55
70 5 -300.00 0.00 -28.55
75 5 -300.00 0.00 -24.35
80 5 -300.00 0.00 -20.01
85 5 -300.00 0.00 -15.63
90 5 -300.00 0.00 -11.28
95 5 -300.00 0.00 -7.06
100 5 -300.00 0.00 -3.05
105 5 -300.00 0.00 0.71
110 5 -300.00 0.00 4.17
115 5 -300.00 0.00 7.31
120 5 -300.00 0.00 10.11
125 5 -300.00 0.00 12.60
130 5 -300.00 0.00 14.77
135 5 -300.00 0.00 16.65
140 5 -300.00 0.00 18.25
145 5 -300.00 0.00 19.59
150 5 -300.00 0.00 20.70
155 5 -300.00 0.00 21.58
160 5 -300.00 0.00 22.25
165 5 -300.00 0.00 22.74
170 5 -300.00 0.00 23.03
175 5 -300.00 0.00 23.15
180 5 -300.00 0.00 23.10
0 10 -300.00 0.00 -64.53
5 10 -300.00 0.00 -64.32
10 10 -300.00 0.00 -63.83
15 10 -300.00 0.00 -63.05
20 10 -300.00 0.00 -61.97
25 10 -300.00 0.00 -60.57
30 10 -300.00 0.00 -58.84
35 10 -300.00 0.00 -56.75
40 10 -300.00 0.00 -54.27
45 10 -300.00 0.00 -51.37
50 10 -300.00 0.00 -48.02
55 10 -300.00 0.00 -44.20
60 10 -300.00 0.00 -39.91
65 10 -300.00 0.00 -35.16
70 10 -300.00 0.00 -30.01
75 10 -300.00 0.00 -24.54
80 10 -300.00 0.00 -18.89
85 10 -300.00 0.00 -13.22
90 10 -300.00 0.00 -7.66
95 10 -300.00 0.00 -2.37
100 10 -300.00 0.00 2.54
105 10 -300.00 0.00 7.02
110 10 -300.00 0.00 11.02
115 10 -300.00 0.00 14.56
120 10 -300.00 0.00 17.64
125 10 -300.00 0.00 20.29
130 10 -300.00 0.00 22.57
135 10 -300.00 0.00 24.49
140 10 -300.00 0.00 26.11
145 10 -300.00 0.00 27.44
150 10 -300.00 0.00 28.52
155 10 -300.00 0.00 29.37
160 10 -300.00 0.00 30.01
165 10 -300.00 0.00 30.45
170 10 -300.00 0.00 30.71
175 10 -300.00 0.00 30.79
180 10 -300.00 0.00 30.71
0 15 -300.00 0.00 -67.78
5 15 -300.00 0.00 -67.56
10 15 -300.00 0.00 -67.05
15 15 -300.00 0.00 -66.24
20 15 -300.00 0.00 -65.13
25 15 -300.00 0.00 -63.69
30 15 -300.00 0.00 -61.90
35 15 -300.00 0.00 -59.72
40 15 -300.00 0.00 -57.12
45 15 -300.00 0.00 -54.05
50 15 -300.00 0.00 -50.47
55 15 -300.00 0.00 -46.35
60 15 -300.00 0.00 -41.67
65 15 -300.00 0.00 -36.45
70 15 -300.00 0.00 -30.73
75 15 -300.00 0.00 -24.64
80 15 -300.00 0.00 -18.34
85 15 -300.00 0.00 -12.03
90 15 -300.00 0.00 -5.90
95 15 -300.00 0.00 -0.13
100 15 -300.00 0.00 5.17
105 15 -300.00 0.00 9.92
110 15 -300.00 0.00 14.11
115 15 -300.00 0.00 17.75
120 15 -300.00 0.00 20.89
125 15 -300.00 0.00 23.58
130 15 -300.00 0.00 25.85
135 15 -300.00 0.00 27.76
140 15 -300.00 0.00 29.35
145 15 -300.00 0.00 30.65
150 15 -300.00 0.00 31.70
155 15 -300.00 0.00 32.52
160 15 -300.00 0.00 33.14
165 15 -300.00 0.00 33.56
170 15 -300.00 0.00 33.80
175 15 -300.00 0.00 33.87
180 15 -300.00 0.00 33.78
0 20 -300.00 0.00 -69.61
5 20 -300.00 0.00 -69.38
10 20 -300.00 0.00 -68.87
15 20 -300.00 0.00 -68.05
20 20 -300.00 0.00 -66.92
25 20 -300.00 0.00 -65.47
30 20 -300.00 0.00 -63.65
35 20 -300.00 0.00 -61.43
40 20 -300.00 0.00 -58.77
45 20 -300.00 0.00 -55.61
50 20 -300.00 0.00 -51.92
55 20 -300.00 0.00 -47.63
60 20 -300.00 0.00 -42.73
65 20 -300.00 0.00 -37.23
70 20 -300.00 0.00 -31.18
75 20 -300.00 0.00 -24.71
80 20 -300.00 0.00 -18.02
85 20 -300.00 0.00 -11.32
90 20 -300.00 0.00 -4.86
95 20 -300.00 0.00 1.18
100 20 -300.00 0.00 6.68
105 20 -300.00 0.00 11.57
110 20 -300.00 0.00 15.84
115 20 -300.00 0.00 19.54
120 20 -300.00 0.00 22.70
125 20 -300.00 0.00 25.38
130 20 -300.00 0.00 27.64
135 20 -300.00 0.00 29.54
140 20 -300.00 0.00 31.11
145 20 -300.00 0.00 32.39
150 20 -300.00 0.00 33.42
155 20 -300.00 0.00 34.23
160 20 -300.00 0.00 34.83
165 20 -300.00 0.00 35.24
170 20 -300.00 0.00 35.47
175 20 -300.00 0.00 35.54
180 20 -300.00 0.00 35.45
0 25 -300.00 0.00 -70.79
5 25 -300.00 0.00 -70.56
10 25 -300.00 0.00 -70.04
15 25 -300.00 0.00 -69.23
20 25 -300.00 0.00 -68.10
25 25 -300.00 0.00 -66.63
30 25 -300.00 0.00 -64.80
35 25 -300.00 0.00 -62.56
40 25 -300.00 0.00 -59.86
45 25 -300.00 0.00 -56.66
50 25 -300.00 0.00 -52.88
55 25 -300.00 0.00 -48.49
60 25 -300.00 0.00 -43.45
65 25 -300.00 0.00 -37.76
70 25 -300.00 0.00 -31.49
75 25 -300.00 0.00 -24.77
80 25 -300.00 0.00 -17.81
85 25 -300.00 0.00 -10.86
90 25 -300.00 0.00 -4.18
95 25 -300.00 0.00 2.03
100 25 -300.00 0.00 7.65
105 25 -300.00 0.00 12.62
110 25 -300.00 0.00 16.95
115 25 -300.00 0.00 20.67
120 25 -300.00 0.00 23.83
125 25 -300.00 0.00 26.51
130 25 -300.00 0.00 28.77
135 25 -300.00 0.00 30.65
140 25 -300.00 0.00 32.21
145 25 -300.00 0.00 33.48
150 25 -300.00 0.00 34.50
155 25 -300.00 0.00 35.30
160 25 -300.00 0.00 35.89
165 25 -300.00 0.00 36.30
170 25 -300.00 0.00 36.53
175 25 -300.00 0.00 36.60
180 25 -300.00 0.00 36.51
0 30 -300.00 0.00 -71.63
5 30 -300.00 0.00 -71.40
10 30 -300.00 0.00 -70.88
15 30 -300.00 0.00 -70.07
20 30 -300.00 0.00 -68.94
25 30 -300.00 0.00 -67.47
30 30 -300.00 0.00 -65.63
35 30 -300.00 0.00 -63.37
40 30 -300.00 0.00 -60.65
45 30 -300.00 0.00 -57.41
50 30 -300.00 0.00 -53.59
55 30 -300.00 0.00 -49.13
60 30 -300.00 0.00 -43.98
65 30 -300.00 0.00 -38.16
70 30 -300.00 0.00 -31.72
75 30 -300.00 0.00 -24.82
80 30 -300.00 0.00 -17.67
85 30 -300.00 0.00 -10.55
90 30 -300.00 0.00 -3.71
95 30 -300.00 0.00 2.62
100 30 -300.00 0.00 8.33
105 30 -300.00 0.00 13.35
110 30 -300.00 0.00 17.70
115 30 -300.00 0.00 21.43
120 30 -300.00 0.00 24.61
125 30 -300.00 0.00 27.28
130 30 -300.00 0.00 29.53
135 30 -300.00 0.00 31.40
140 30 -300.00 0.00 32.96
145 30 -300.00 0.00 34.22
150 30 -300.00 0.00 35.24
155 30 -300.00 0.00 36.03
160 30 -300.00 0.00 36.62
165 30 -300.00 0.00 37.03
170 30 -300.00 0.00 37.27
175 30 -300.00 0.00 37.34
180 30 -300.00 0.00 37.25
0 35 -300.00 0.00 -72.26
5 35 -300.00 0.00 -72.04
10 35 -300.00 0.00 -71.52
15 35 -300.00 0.00 -70.71
20 35 -300.00 0.00 -69.58
25 35 -300.00 0.00 -68.11
30 35 -300.00 0.00 -66.26
35 35 -300.00 0.00 -64.00
40 35 -300.00 0.00 -61.27
45 35 -300.00 0.00 -58.00
50 35 -300.00 0.00 -54.14
55 35 -300.00 0.00 -49.62
60 35 -300.00 0.00 -44.40
65 35 -300.00 0.00 -38.47
70 35 -300.00 0.00 -31.91
75 35 -300.00 0.00 -24.87
80 35 -300.00 0.00 -17.58
85 35 -300.00 0.00 -10.32
90 35 -300.00 0.00 -3.38
95 35 -300.00 0.00 3.04
100 35 -300.00 0.00 8.81
105 35 -300.00 0.00 13.87
110 35 -300.00 0.00 18.24
115 35 -300.00 0.00 21.98
120 35 -300.00 0.00 25.15
125 35 -300.00 0.00 27.83
130 35 -300.00 0.00 30.07
135 35 -300.00 0.00 31.94
140 35 -300.00 0.00 33.49
145 35 -300.00 0.00 34.76
150 35 -300.00 0.00 35.77
155 35 -300.00 0.00 36.57
160 35 -300.00 0.00 37.16
165 35 -300.00 0.00 37.57
170 35 -300.00 0.00 37.81
175 35 -300.00 0.00 37.89
180 35 -300.00 0.00 37.81
0 40 -300.00 0.00 -72.77
... 159721 more lines ...
|
| Attachment 4: XF_model_19_96.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -83.71 0.00 -158.06
5 0 -17.86 0.02 -153.75
10 0 -11.87 0.06 -148.66
15 0 -8.40 0.14 -144.50
20 0 -5.97 0.25 -140.71
25 0 -4.13 0.39 -136.31
30 0 -2.66 0.54 -131.91
35 0 -1.45 0.72 -127.38
40 0 -0.45 0.90 -123.42
45 0 0.39 1.09 -119.24
50 0 1.09 1.29 -115.23
55 0 1.69 1.47 -111.53
60 0 2.18 1.65 -107.68
65 0 2.59 1.81 -104.22
70 0 2.91 1.95 -100.95
75 0 3.15 2.07 -97.77
80 0 3.33 2.15 -94.90
85 0 3.43 2.20 -92.40
90 0 3.46 2.22 -89.68
95 0 3.43 2.20 -87.43
100 0 3.33 2.15 -85.51
105 0 3.15 2.07 -84.01
110 0 2.91 1.95 -82.20
115 0 2.59 1.81 -80.97
120 0 2.18 1.65 -79.70
125 0 1.69 1.47 -78.83
130 0 1.09 1.29 -77.85
135 0 0.39 1.09 -77.23
140 0 -0.45 0.90 -77.60
145 0 -1.45 0.72 -77.60
150 0 -2.66 0.54 -78.04
155 0 -4.13 0.39 -78.59
160 0 -5.97 0.25 -79.94
165 0 -8.40 0.14 -81.34
170 0 -11.87 0.07 -84.60
175 0 -17.86 0.02 -89.05
180 0 -83.70 0.00 -95.44
0 5 -83.74 0.00 -20.42
5 5 -17.86 0.02 -21.56
10 5 -11.87 0.06 -22.65
15 5 -8.40 0.14 -23.78
20 5 -5.97 0.25 -24.81
25 5 -4.13 0.39 -25.80
30 5 -2.66 0.54 -26.78
35 5 -1.45 0.72 -27.69
40 5 -0.45 0.90 -28.54
45 5 0.39 1.09 -29.38
50 5 1.09 1.29 -30.15
55 5 1.69 1.47 -30.88
60 5 2.18 1.65 -31.59
65 5 2.59 1.81 -32.25
70 5 2.91 1.95 -32.87
75 5 3.15 2.07 -33.45
80 5 3.33 2.15 -33.97
85 5 3.43 2.20 -34.45
90 5 3.46 2.22 -34.85
95 5 3.43 2.20 -35.18
100 5 3.33 2.15 -35.40
105 5 3.15 2.07 -35.52
110 5 2.91 1.95 -35.51
115 5 2.59 1.81 -35.38
120 5 2.18 1.65 -35.10
125 5 1.69 1.47 -34.70
130 5 1.09 1.29 -34.17
135 5 0.39 1.09 -33.48
140 5 -0.45 0.90 -32.71
145 5 -1.45 0.72 -31.84
150 5 -2.66 0.54 -30.83
155 5 -4.13 0.39 -29.77
160 5 -5.97 0.25 -28.61
165 5 -8.40 0.14 -27.42
170 5 -11.87 0.07 -26.17
175 5 -17.86 0.02 -24.90
180 5 -83.74 0.00 -23.60
0 10 -83.84 0.00 -19.91
5 10 -17.86 0.02 -20.98
10 10 -11.87 0.06 -22.04
15 10 -8.40 0.14 -23.08
20 10 -5.97 0.25 -24.09
25 10 -4.13 0.39 -25.07
30 10 -2.66 0.54 -26.00
35 10 -1.45 0.72 -26.89
40 10 -0.45 0.90 -27.75
45 10 0.39 1.09 -28.55
50 10 1.09 1.29 -29.32
55 10 1.69 1.47 -30.06
60 10 2.18 1.65 -30.76
65 10 2.59 1.81 -31.42
70 10 2.91 1.95 -32.05
75 10 3.15 2.07 -32.64
80 10 3.33 2.15 -33.17
85 10 3.43 2.20 -33.66
90 10 3.46 2.22 -34.07
95 10 3.43 2.20 -34.41
100 10 3.33 2.15 -34.64
105 10 3.15 2.07 -34.77
110 10 2.91 1.95 -34.79
115 10 2.59 1.81 -34.68
120 10 2.18 1.65 -34.44
125 10 1.69 1.47 -34.06
130 10 1.09 1.29 -33.55
135 10 0.39 1.09 -32.91
140 10 -0.45 0.90 -32.16
145 10 -1.45 0.72 -31.30
150 10 -2.66 0.54 -30.34
155 10 -4.13 0.39 -29.31
160 10 -5.97 0.25 -28.20
165 10 -8.40 0.14 -27.05
170 10 -11.87 0.07 -25.86
175 10 -17.86 0.02 -24.64
180 10 -83.84 0.00 -23.37
0 15 -84.01 0.00 -19.75
5 15 -17.86 0.02 -20.78
10 15 -11.87 0.06 -21.80
15 15 -8.40 0.14 -22.82
20 15 -5.97 0.25 -23.80
25 15 -4.13 0.39 -24.75
30 15 -2.66 0.54 -25.67
35 15 -1.45 0.72 -26.54
40 15 -0.45 0.90 -27.38
45 15 0.39 1.09 -28.17
50 15 1.09 1.29 -28.93
55 15 1.69 1.47 -29.66
60 15 2.18 1.65 -30.36
65 15 2.59 1.81 -31.01
70 15 2.91 1.95 -31.63
75 15 3.15 2.07 -32.21
80 15 3.33 2.15 -32.75
85 15 3.43 2.20 -33.22
90 15 3.46 2.22 -33.63
95 15 3.43 2.20 -33.96
100 15 3.33 2.15 -34.19
105 15 3.15 2.07 -34.33
110 15 2.91 1.95 -34.35
115 15 2.59 1.81 -34.24
120 15 2.18 1.65 -34.00
125 15 1.69 1.47 -33.65
130 15 1.09 1.29 -33.16
135 15 0.39 1.09 -32.54
140 15 -0.45 0.90 -31.82
145 15 -1.45 0.72 -30.99
150 15 -2.66 0.54 -30.07
155 15 -4.13 0.39 -29.06
160 15 -5.97 0.25 -28.00
165 15 -8.40 0.14 -26.87
170 15 -11.87 0.07 -25.71
175 15 -17.86 0.02 -24.52
180 15 -84.01 0.00 -23.30
0 20 -84.25 0.00 -19.66
5 20 -17.86 0.02 -20.66
10 20 -11.87 0.06 -21.65
15 20 -8.40 0.14 -22.63
20 20 -5.97 0.25 -23.59
25 20 -4.13 0.39 -24.52
30 20 -2.66 0.54 -25.42
35 20 -1.45 0.72 -26.27
40 20 -0.45 0.90 -27.10
45 20 0.39 1.09 -27.88
50 20 1.09 1.29 -28.63
55 20 1.69 1.47 -29.35
60 20 2.18 1.65 -30.03
65 20 2.59 1.81 -30.67
70 20 2.91 1.95 -31.28
75 20 3.15 2.07 -31.84
80 20 3.33 2.15 -32.36
85 20 3.43 2.20 -32.82
90 20 3.46 2.22 -33.21
95 20 3.43 2.20 -33.52
100 20 3.33 2.15 -33.75
105 20 3.15 2.07 -33.88
110 20 2.91 1.95 -33.89
115 20 2.59 1.81 -33.79
120 20 2.18 1.65 -33.57
125 20 1.69 1.47 -33.22
130 20 1.09 1.29 -32.76
135 20 0.39 1.09 -32.17
140 20 -0.45 0.90 -31.48
145 20 -1.45 0.72 -30.68
150 20 -2.66 0.54 -29.80
155 20 -4.13 0.39 -28.83
160 20 -5.97 0.25 -27.80
165 20 -8.40 0.14 -26.72
170 20 -11.87 0.07 -25.60
175 20 -17.86 0.02 -24.44
180 20 -84.25 0.00 -23.27
0 25 -84.56 0.00 -19.61
5 25 -17.86 0.02 -20.56
10 25 -11.87 0.06 -21.52
15 25 -8.40 0.14 -22.47
20 25 -5.97 0.25 -23.40
25 25 -4.13 0.39 -24.30
30 25 -2.66 0.54 -25.17
35 25 -1.45 0.72 -26.01
40 25 -0.45 0.90 -26.81
45 25 0.39 1.09 -27.58
50 25 1.09 1.29 -28.32
55 25 1.69 1.47 -29.01
60 25 2.18 1.65 -29.68
65 25 2.59 1.81 -30.31
70 25 2.91 1.95 -30.89
75 25 3.15 2.07 -31.44
80 25 3.33 2.15 -31.93
85 25 3.43 2.20 -32.37
90 25 3.46 2.22 -32.75
95 25 3.43 2.20 -33.04
100 25 3.33 2.15 -33.25
105 25 3.15 2.07 -33.37
110 25 2.91 1.95 -33.38
115 25 2.59 1.81 -33.28
120 25 2.18 1.65 -33.07
125 25 1.69 1.47 -32.74
130 25 1.09 1.29 -32.30
135 25 0.39 1.09 -31.75
140 25 -0.45 0.90 -31.09
145 25 -1.45 0.72 -30.35
150 25 -2.66 0.54 -29.50
155 25 -4.13 0.39 -28.58
160 25 -5.97 0.25 -27.60
165 25 -8.40 0.14 -26.56
170 25 -11.87 0.07 -25.49
175 25 -17.86 0.02 -24.38
180 25 -84.56 0.00 -23.25
0 30 -84.95 0.00 -19.57
5 30 -17.86 0.02 -20.48
10 30 -11.87 0.06 -21.40
15 30 -8.40 0.14 -22.31
20 30 -5.97 0.25 -23.20
25 30 -4.13 0.39 -24.07
30 30 -2.66 0.54 -24.92
35 30 -1.45 0.72 -25.73
40 30 -0.45 0.90 -26.51
45 30 0.39 1.09 -27.26
50 30 1.09 1.29 -27.97
55 30 1.69 1.47 -28.65
60 30 2.18 1.65 -29.30
65 30 2.59 1.81 -29.90
70 30 2.91 1.95 -30.47
75 30 3.15 2.07 -30.99
80 30 3.33 2.15 -31.45
85 30 3.43 2.20 -31.87
90 30 3.46 2.22 -32.21
95 30 3.43 2.20 -32.49
100 30 3.33 2.15 -32.69
105 30 3.15 2.07 -32.79
110 30 2.91 1.95 -32.80
115 30 2.59 1.81 -32.71
120 30 2.18 1.65 -32.51
125 30 1.69 1.47 -32.20
130 30 1.09 1.29 -31.79
135 30 0.39 1.09 -31.27
140 30 -0.45 0.90 -30.66
145 30 -1.45 0.72 -29.95
150 30 -2.66 0.54 -29.16
155 30 -4.13 0.39 -28.30
160 30 -5.97 0.25 -27.37
165 30 -8.40 0.14 -26.39
170 30 -11.87 0.07 -25.36
175 30 -17.86 0.02 -24.31
180 30 -84.96 0.00 -23.23
0 35 -85.43 0.00 -19.55
5 35 -17.86 0.02 -20.41
10 35 -11.87 0.06 -21.27
15 35 -8.40 0.14 -22.14
20 35 -5.97 0.25 -22.99
25 35 -4.13 0.39 -23.82
30 35 -2.66 0.54 -24.63
35 35 -1.45 0.72 -25.42
40 35 -0.45 0.90 -26.17
45 35 0.39 1.09 -26.90
50 35 1.09 1.29 -27.58
55 35 1.69 1.47 -28.24
60 35 2.18 1.65 -28.86
65 35 2.59 1.81 -29.44
70 35 2.91 1.95 -29.98
75 35 3.15 2.07 -30.47
80 35 3.33 2.15 -30.91
85 35 3.43 2.20 -31.30
90 35 3.46 2.22 -31.61
95 35 3.43 2.20 -31.87
100 35 3.33 2.15 -32.05
105 35 3.15 2.07 -32.14
110 35 2.91 1.95 -32.15
115 35 2.59 1.81 -32.06
120 35 2.18 1.65 -31.88
125 35 1.69 1.47 -31.60
130 35 1.09 1.29 -31.22
135 35 0.39 1.09 -30.74
140 35 -0.45 0.90 -30.17
145 35 -1.45 0.72 -29.52
150 35 -2.66 0.54 -28.79
155 35 -4.13 0.39 -27.98
160 35 -5.97 0.25 -27.11
165 35 -8.40 0.14 -26.19
170 35 -11.87 0.07 -25.23
175 35 -17.86 0.02 -24.23
180 35 -85.44 0.00 -23.22
0 40 -86.01 0.00 -19.53
... 159721 more lines ...
|
| Attachment 5: XF_model_29_87.txt
|
freq : 83.33 MHz
SWR : 1.965000
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -77.59 0.00 -23.13
5 0 -17.30 0.02 -23.13
10 0 -11.30 0.07 -23.11
15 0 -7.83 0.16 -23.07
20 0 -5.40 0.29 -23.00
25 0 -3.56 0.44 -22.93
30 0 -2.09 0.62 -22.84
35 0 -0.89 0.82 -22.73
40 0 0.11 1.03 -22.61
45 0 0.95 1.24 -22.47
50 0 1.66 1.46 -22.33
55 0 2.25 1.68 -22.17
60 0 2.74 1.88 -22.01
65 0 3.14 2.06 -21.84
70 0 3.46 2.22 -21.66
75 0 3.71 2.35 -21.48
80 0 3.88 2.44 -21.29
85 0 3.99 2.50 -21.11
90 0 4.02 2.52 -20.92
95 0 3.99 2.50 -20.74
100 0 3.88 2.45 -20.55
105 0 3.71 2.35 -20.38
110 0 3.47 2.22 -20.20
115 0 3.15 2.06 -20.03
120 0 2.74 1.88 -19.87
125 0 2.25 1.68 -19.72
130 0 1.66 1.47 -19.58
135 0 0.96 1.25 -19.44
140 0 0.12 1.03 -19.31
145 0 -0.88 0.82 -19.20
150 0 -2.08 0.62 -19.10
155 0 -3.55 0.44 -19.02
160 0 -5.39 0.29 -18.94
165 0 -7.82 0.17 -18.89
170 0 -11.29 0.07 -18.84
175 0 -17.28 0.02 -18.82
180 0 -77.55 0.00 -18.81
0 5 -77.73 0.00 -18.36
5 5 -17.30 0.02 -18.44
10 5 -11.30 0.07 -18.54
15 5 -7.83 0.16 -18.64
20 5 -5.40 0.29 -18.77
25 5 -3.56 0.44 -18.92
30 5 -2.09 0.62 -19.09
35 5 -0.89 0.82 -19.27
40 5 0.11 1.03 -19.47
45 5 0.95 1.24 -19.67
50 5 1.66 1.46 -19.89
55 5 2.25 1.68 -20.12
60 5 2.74 1.88 -20.35
65 5 3.14 2.06 -20.60
70 5 3.46 2.22 -20.84
75 5 3.71 2.35 -21.08
80 5 3.88 2.44 -21.33
85 5 3.99 2.50 -21.57
90 5 4.02 2.52 -21.81
95 5 3.99 2.50 -22.04
100 5 3.88 2.45 -22.28
105 5 3.71 2.35 -22.50
110 5 3.47 2.22 -22.72
115 5 3.15 2.06 -22.92
120 5 2.74 1.88 -23.12
125 5 2.25 1.68 -23.30
130 5 1.66 1.47 -23.47
135 5 0.96 1.25 -23.62
140 5 0.12 1.03 -23.75
145 5 -0.88 0.82 -23.85
150 5 -2.08 0.62 -23.94
155 5 -3.55 0.44 -24.00
160 5 -5.39 0.29 -24.04
165 5 -7.82 0.17 -24.06
170 5 -11.29 0.07 -24.04
175 5 -17.28 0.02 -24.01
180 5 -77.69 0.00 -23.95
0 10 -77.94 0.00 -16.18
5 10 -17.30 0.02 -16.28
10 10 -11.30 0.07 -16.44
15 10 -7.83 0.16 -16.61
20 10 -5.40 0.29 -16.84
25 10 -3.56 0.44 -17.09
30 10 -2.09 0.62 -17.37
35 10 -0.89 0.82 -17.69
40 10 0.11 1.03 -18.03
45 10 0.95 1.24 -18.40
50 10 1.66 1.46 -18.78
55 10 2.25 1.68 -19.19
60 10 2.74 1.88 -19.60
65 10 3.14 2.06 -20.03
70 10 3.46 2.22 -20.46
75 10 3.71 2.35 -20.89
80 10 3.88 2.44 -21.33
85 10 3.99 2.50 -21.76
90 10 4.02 2.52 -22.19
95 10 3.99 2.50 -22.61
100 10 3.88 2.45 -23.02
105 10 3.71 2.35 -23.43
110 10 3.47 2.22 -23.82
115 10 3.15 2.06 -24.20
120 10 2.74 1.88 -24.55
125 10 2.25 1.68 -24.88
130 10 1.66 1.47 -25.19
135 10 0.96 1.25 -25.46
140 10 0.12 1.03 -25.71
145 10 -0.88 0.82 -25.92
150 10 -2.08 0.62 -26.09
155 10 -3.55 0.44 -26.22
160 10 -5.39 0.29 -26.30
165 10 -7.82 0.17 -26.35
170 10 -11.29 0.07 -26.36
175 10 -17.28 0.02 -26.33
180 10 -77.89 0.00 -26.25
0 15 -78.23 0.00 -14.91
5 15 -17.30 0.02 -15.04
10 15 -11.30 0.07 -15.22
15 15 -7.83 0.16 -15.44
20 15 -5.40 0.29 -15.71
25 15 -3.56 0.44 -16.02
30 15 -2.09 0.62 -16.37
35 15 -0.89 0.82 -16.76
40 15 0.11 1.03 -17.19
45 15 0.95 1.24 -17.64
50 15 1.66 1.46 -18.12
55 15 2.25 1.68 -18.63
60 15 2.74 1.88 -19.14
65 15 3.14 2.06 -19.68
70 15 3.46 2.22 -20.21
75 15 3.71 2.35 -20.76
80 15 3.88 2.44 -21.30
85 15 3.99 2.50 -21.85
90 15 4.02 2.52 -22.39
95 15 3.99 2.50 -22.92
100 15 3.88 2.45 -23.44
105 15 3.71 2.35 -23.94
110 15 3.47 2.22 -24.43
115 15 3.15 2.06 -24.91
120 15 2.74 1.88 -25.35
125 15 2.25 1.68 -25.77
130 15 1.66 1.47 -26.16
135 15 0.96 1.25 -26.51
140 15 0.12 1.03 -26.82
145 15 -0.88 0.82 -27.09
150 15 -2.08 0.62 -27.31
155 15 -3.55 0.44 -27.48
160 15 -5.39 0.29 -27.60
165 15 -7.82 0.17 -27.67
170 15 -11.29 0.07 -27.68
175 15 -17.28 0.02 -27.65
180 15 -78.17 0.00 -27.56
0 20 -78.59 0.00 -14.07
5 20 -17.30 0.02 -14.21
10 20 -11.30 0.07 -14.41
15 20 -7.83 0.16 -14.66
20 20 -5.40 0.29 -14.95
25 20 -3.56 0.44 -15.30
30 20 -2.09 0.62 -15.70
35 20 -0.89 0.82 -16.14
40 20 0.11 1.03 -16.61
45 20 0.95 1.24 -17.12
50 20 1.66 1.46 -17.66
55 20 2.25 1.68 -18.23
60 20 2.74 1.88 -18.82
65 20 3.14 2.06 -19.42
70 20 3.46 2.22 -20.03
75 20 3.71 2.35 -20.64
80 20 3.88 2.44 -21.26
85 20 3.99 2.50 -21.88
90 20 4.02 2.52 -22.49
95 20 3.99 2.50 -23.09
100 20 3.88 2.45 -23.69
105 20 3.71 2.35 -24.26
110 20 3.47 2.22 -24.82
115 20 3.15 2.06 -25.36
120 20 2.74 1.88 -25.87
125 20 2.25 1.68 -26.34
130 20 1.66 1.47 -26.78
135 20 0.96 1.25 -27.18
140 20 0.12 1.03 -27.53
145 20 -0.88 0.82 -27.84
150 20 -2.08 0.62 -28.09
155 20 -3.55 0.44 -28.29
160 20 -5.40 0.29 -28.43
165 20 -7.82 0.17 -28.52
170 20 -11.29 0.07 -28.54
175 20 -17.28 0.02 -28.51
180 20 -78.53 0.00 -28.42
0 25 -79.03 0.00 -13.47
5 25 -17.30 0.02 -13.62
10 25 -11.30 0.07 -13.83
15 25 -7.83 0.16 -14.09
20 25 -5.40 0.29 -14.41
25 25 -3.56 0.44 -14.78
30 25 -2.09 0.62 -15.20
35 25 -0.89 0.82 -15.67
40 25 0.11 1.03 -16.18
45 25 0.95 1.24 -16.73
50 25 1.66 1.46 -17.31
55 25 2.25 1.68 -17.92
60 25 2.74 1.88 -18.55
65 25 3.14 2.06 -19.20
70 25 3.46 2.22 -19.86
75 25 3.71 2.35 -20.53
80 25 3.88 2.44 -21.20
85 25 3.99 2.50 -21.87
90 25 4.02 2.52 -22.54
95 25 3.99 2.50 -23.20
100 25 3.88 2.45 -23.84
105 25 3.71 2.35 -24.47
110 25 3.47 2.22 -25.08
115 25 3.15 2.06 -25.66
120 25 2.74 1.88 -26.21
125 25 2.25 1.68 -26.73
130 25 1.66 1.47 -27.21
135 25 0.96 1.25 -27.64
140 25 0.12 1.03 -28.03
145 25 -0.88 0.82 -28.36
150 25 -2.08 0.62 -28.64
155 25 -3.55 0.44 -28.86
160 25 -5.40 0.29 -29.02
165 25 -7.82 0.17 -29.12
170 25 -11.29 0.07 -29.15
175 25 -17.28 0.02 -29.13
180 25 -78.97 0.00 -29.04
0 30 -79.57 0.00 -13.01
5 30 -17.30 0.02 -13.16
10 30 -11.30 0.07 -13.37
15 30 -7.83 0.16 -13.65
20 30 -5.40 0.29 -13.97
25 30 -3.56 0.44 -14.36
30 30 -2.09 0.62 -14.80
35 30 -0.89 0.82 -15.29
40 30 0.11 1.03 -15.82
45 30 0.95 1.24 -16.40
50 30 1.66 1.46 -17.01
55 30 2.25 1.68 -17.65
60 30 2.74 1.88 -18.32
65 30 3.14 2.06 -19.00
70 30 3.46 2.22 -19.70
75 30 3.71 2.35 -20.41
80 30 3.88 2.44 -21.12
85 30 3.99 2.50 -21.84
90 30 4.02 2.52 -22.55
95 30 3.99 2.50 -23.24
100 30 3.88 2.45 -23.93
105 30 3.71 2.35 -24.60
110 30 3.47 2.22 -25.25
115 30 3.15 2.06 -25.87
120 30 2.74 1.88 -26.46
125 30 2.25 1.68 -27.01
130 30 1.66 1.47 -27.52
135 30 0.96 1.25 -27.98
140 30 0.12 1.03 -28.39
145 30 -0.88 0.82 -28.75
150 30 -2.08 0.62 -29.05
155 30 -3.55 0.44 -29.28
160 30 -5.40 0.29 -29.46
165 30 -7.82 0.17 -29.57
170 30 -11.29 0.07 -29.61
175 30 -17.28 0.02 -29.59
180 30 -79.50 0.00 -29.50
0 35 -80.22 0.00 -12.65
5 35 -17.30 0.02 -12.80
10 35 -11.30 0.07 -13.01
15 35 -7.83 0.16 -13.28
20 35 -5.40 0.29 -13.62
25 35 -3.56 0.44 -14.02
30 35 -2.09 0.62 -14.47
35 35 -0.89 0.82 -14.97
40 35 0.11 1.03 -15.52
45 35 0.95 1.24 -16.11
50 35 1.66 1.46 -16.75
55 35 2.25 1.68 -17.41
60 35 2.74 1.88 -18.10
65 35 3.14 2.06 -18.81
70 35 3.46 2.22 -19.54
75 35 3.71 2.35 -20.28
80 35 3.88 2.44 -21.03
85 35 3.99 2.50 -21.78
90 35 4.02 2.52 -22.52
95 35 3.99 2.50 -23.26
100 35 3.88 2.45 -23.98
105 35 3.71 2.35 -24.68
110 35 3.47 2.22 -25.36
115 35 3.15 2.06 -26.02
120 35 2.74 1.88 -26.63
125 35 2.25 1.68 -27.21
130 35 1.66 1.47 -27.75
135 35 0.96 1.25 -28.23
140 35 0.12 1.03 -28.66
145 35 -0.88 0.82 -29.04
150 35 -2.08 0.62 -29.36
155 35 -3.55 0.44 -29.61
160 35 -5.40 0.29 -29.80
165 35 -7.82 0.17 -29.92
170 35 -11.29 0.07 -29.97
175 35 -17.28 0.02 -29.96
180 35 -80.14 0.00 -29.87
0 40 -80.99 0.00 -12.34
... 159721 more lines ...
|
|
|
2
|
Wed Feb 6 17:28:29 2019 |
Julie Rolla | Bicone User Manual (w/ links to Dipole work) | Hi all,
The working copy of the user manual can be found here: https://www.overleaf.com/6955763886pjtkckprxsyp
It still need to be tested by someone new to see if edits need to be made. I have uploaded the copy that is finalized as of this day. Note if you click on the overleaf version that may be more up-to-date. This copy is added as a PDF so we can keep track of versions. |
| Attachment 1: GENETIS_Manual_1_25_19.pdf
|
|
|
17
|
Thu Nov 7 16:55:00 2019 |
Cade Sbrocco | Archiving Old Genetis Manual | |
| Attachment 1: GENETIS_Manual.pdf
|
|
|
11
|
Wed Apr 3 16:38:07 2019 |
Julie Rolla | AraSim update | General update
The following were the fixes we made to make AraSim run:
(1) As of March, the loop breaks at AraSim. We have found that our program XFintoARA.py does not properly format the output in the form that AraSim is looking for (as an input). Jorge gave us a sample AraSim input file to help us edit XFintoARA.py so that it formats the file correctly. Note that we found white space is also must match in order for the format to be correct.
Now that we have fixed this, AraSim now runs. Attached is the following sample Jorge gave us of the proper AraSim inputs. If needed, it's available to refer back to.
(2) After correcting the AraSim input file formats (outputs of XFintoARA.py), we ran it. It failed to finish running after hours of run time. We found that we were using default input parameters for AraSim -- which wasn't triggering in a timely fashion. Once we edited the parameters, it ran successfully. Note that this was not run within the loop. Confirmation of the full loop running has not been completed.
The parameters of interest need to be edited in setup.txt. To see which file in AraSim I am referring to, please see the .txt file attached. Note that we will be editing the bash script to allow the user to change these input parameters there. The following red lines are what need to be set:
EXPONENT=21
NNU=100 // number of neutrino events
NNU_PASSED=10 // number of neutrino events that are allowed to pass the trigger
ONLY_PASSED_EVENTS=1 // 0 (default): AraSim throws NNU events whether or not they pass; 1: AraSim throws events until the number of events that pass the trigger is equal to NNU_PASSED (WARNING: may cause long run times if reasonable values are not chosen)
NOISE_WAVEFORM_GENERATE_MODE=0 // generate new noise waveforms for each events
NOISE_EVENTS=16 // number of pure noise waveforms
TRIG_ANALYSIS_MODE=0 // 0 = signal + noise, 1 = signal only, 2 = noise only
DETECTOR=1 // ARA stations 1 to 7
NOFZ=1
core_x=10000
core_y=10000
EXPONENT gives the energy of the neutrinos. Note that higher energies will have more pass, as the energies don't dissipate as rapidly. For now, we are running at this energy. Eventally, we will want to use the full Kotera spectrum. Otherwsie, we would be finding the performance of an antenna at only one energy. Just to check the loop, we are going to use EXPONENT=21 to get events to trigger faster.
ONLY_PASSED_EVENTS decides which of the two options to use to determine when AraSim is finished running: (1) Throws 'NNU' number of events at antennas and stops regardless of how many pass (in this case ONLY_PASSED_EVENTS=0) , or (2) throws as many events at the antenna as needed until 'NNU_PASSED' number triggers (in this case ONLY_PASSED_EVENTS=1). Ie if this number is 10, we throw as many neutrinos at the antenna as necessary until 10 events trigger. Note that this can result in a longer run time. Additionally, a higher EXPONENT will make them trigger faster.
TRIG_ANALYSIS_MODE is a setting that determines what is being thrown. Ie- 0=signal + noise (which is what we want, and is the 'real' case), 1=signal only (would make it run faster), 2=noise only (not of interest for us).
To-Do List
(1) Commit version of XFintoARA.py since recent formatting change.
(2) Figure out how to add in Kotera spectrum (note that we wont want to do this until the WHOLE loop is functional)
(3) Edit the bash script to recreat the setup.txt making the variables in red above an editable option in the top of the bash script. Ie our variables in red above will be at the top of the bash script. Later in the bash script we will need to have it create this .txt file with the user inputs for those variables. We can do this by either (1) creating a skeleton, or (2) creating another .sh script that is run in our XFLoop.sh -- which writes the first handfull of lines based on the user inputs for our variables in red above, and then pastes in the rest of what is unchanged in setup.txt.
(4) Run the whole loop: update- we tried this today and keep getting a Veff-0 for our first antenna along with the error
Warning in <TFile::Init>: no StreamerInfo found in /datapool/software/anita/lib/G__RootFftwWrapper_rdict.pcm therefore preventing schema evolution when reading this file.
Error in <TStreamerInfo::Build>: AraEventCalibrator, discarding: unsigned short* fAtriPeds, no [dimension]
others run as expected
(5) Work on the paper: Amy added a skeleton of sections to add for the GA. Date of completion should be in-line with APS.
(6) APS talk slides.
(7) Edit the manual and add:
- The changes that will occur when task (3) is done.
- A sample of how AraSim inputs -- or XFintoARA.py outputs -- should look.
- This stuff: http://radiorm.physics.ohio-state.edu/elog/GENETIS/8
- Max's AraSim plotting software (when it's done)
- Evelyn and Sophie's plotting software info (when it's done).
(8) Finish Max's AraSim plotting software.
(9) Finish Evenlyn and Sophie's plotting softare. |
| Attachment 1: ARA_bicone6in_output.txt
|
freq : 83.33 MHz
SWR : 2.18
Theta Phi Gain(dB) Gain Phase(deg)
0 0 -80.83 0.00 -0.41
5 0 -21.10 0.01 168.18
10 0 -15.10 0.03 168.17
15 0 -11.62 0.07 168.14
20 0 -9.19 0.12 168.09
25 0 -7.34 0.18 168.04
30 0 -5.86 0.26 167.97
35 0 -4.65 0.34 167.91
40 0 -3.64 0.43 167.83
45 0 -2.79 0.53 167.76
50 0 -2.08 0.62 167.70
55 0 -1.48 0.71 167.64
60 0 -0.98 0.80 167.59
65 0 -0.57 0.88 167.55
70 0 -0.25 0.94 167.53
75 0 0.00 1.00 167.51
80 0 0.18 1.04 167.50
85 0 0.29 1.07 167.50
90 0 0.32 1.08 167.50
95 0 0.29 1.07 167.50
100 0 0.18 1.04 167.50
105 0 0.01 1.00 167.51
110 0 -0.25 0.94 167.53
115 0 -0.57 0.88 167.55
120 0 -0.98 0.80 167.59
125 0 -1.48 0.71 167.64
130 0 -2.08 0.62 167.70
135 0 -2.79 0.53 167.77
140 0 -3.64 0.43 167.84
145 0 -4.65 0.34 167.91
150 0 -5.86 0.26 167.98
155 0 -7.33 0.18 168.04
160 0 -9.19 0.12 168.10
165 0 -11.62 0.07 168.15
170 0 -15.09 0.03 168.18
175 0 -21.08 0.01 168.21
180 0 -80.83 0.00 179.59
0 5 -80.83 0.00 -0.41
5 5 -21.10 0.01 168.18
10 5 -15.10 0.03 168.17
15 5 -11.62 0.07 168.14
20 5 -9.19 0.12 168.09
25 5 -7.34 0.18 168.04
30 5 -5.86 0.26 167.97
35 5 -4.65 0.34 167.91
40 5 -3.64 0.43 167.83
45 5 -2.79 0.53 167.76
50 5 -2.08 0.62 167.70
55 5 -1.48 0.71 167.64
60 5 -0.98 0.80 167.59
65 5 -0.57 0.88 167.55
70 5 -0.25 0.94 167.53
75 5 0.00 1.00 167.51
80 5 0.18 1.04 167.50
85 5 0.29 1.07 167.50
90 5 0.32 1.08 167.50
95 5 0.29 1.07 167.50
100 5 0.18 1.04 167.50
105 5 0.01 1.00 167.51
110 5 -0.25 0.94 167.53
115 5 -0.57 0.88 167.56
120 5 -0.98 0.80 167.59
125 5 -1.48 0.71 167.64
130 5 -2.08 0.62 167.70
135 5 -2.79 0.53 167.77
140 5 -3.64 0.43 167.84
145 5 -4.65 0.34 167.91
150 5 -5.86 0.26 167.98
155 5 -7.33 0.18 168.04
160 5 -9.19 0.12 168.10
165 5 -11.62 0.07 168.15
170 5 -15.09 0.03 168.18
175 5 -21.08 0.01 168.21
180 5 -80.83 0.00 179.59
0 10 -80.83 0.00 -0.41
5 10 -21.10 0.01 168.18
10 10 -15.10 0.03 168.17
15 10 -11.62 0.07 168.14
20 10 -9.19 0.12 168.09
25 10 -7.34 0.18 168.04
30 10 -5.86 0.26 167.97
35 10 -4.65 0.34 167.91
40 10 -3.64 0.43 167.83
45 10 -2.79 0.53 167.76
50 10 -2.08 0.62 167.70
55 10 -1.48 0.71 167.64
60 10 -0.98 0.80 167.59
65 10 -0.57 0.88 167.55
70 10 -0.25 0.94 167.53
75 10 0.01 1.00 167.51
80 10 0.18 1.04 167.50
85 10 0.29 1.07 167.50
90 10 0.32 1.08 167.50
95 10 0.29 1.07 167.50
100 10 0.18 1.04 167.50
105 10 0.01 1.00 167.51
110 10 -0.24 0.95 167.53
115 10 -0.57 0.88 167.56
120 10 -0.98 0.80 167.59
125 10 -1.48 0.71 167.64
130 10 -2.08 0.62 167.70
135 10 -2.79 0.53 167.77
140 10 -3.64 0.43 167.84
145 10 -4.65 0.34 167.91
150 10 -5.86 0.26 167.98
155 10 -7.33 0.18 168.04
160 10 -9.19 0.12 168.10
165 10 -11.62 0.07 168.15
170 10 -15.09 0.03 168.18
175 10 -21.08 0.01 168.21
180 10 -80.83 0.00 179.59
0 15 -80.83 0.00 -0.41
5 15 -21.10 0.01 168.18
10 15 -15.10 0.03 168.17
15 15 -11.62 0.07 168.14
20 15 -9.19 0.12 168.09
25 15 -7.34 0.18 168.04
30 15 -5.86 0.26 167.97
35 15 -4.65 0.34 167.91
40 15 -3.64 0.43 167.83
45 15 -2.79 0.53 167.76
50 15 -2.08 0.62 167.70
55 15 -1.48 0.71 167.64
60 15 -0.98 0.80 167.59
65 15 -0.57 0.88 167.55
70 15 -0.25 0.94 167.53
75 15 0.01 1.00 167.51
80 15 0.18 1.04 167.50
85 15 0.29 1.07 167.50
90 15 0.32 1.08 167.50
95 15 0.29 1.07 167.50
100 15 0.18 1.04 167.50
105 15 0.01 1.00 167.51
110 15 -0.24 0.95 167.53
115 15 -0.57 0.88 167.56
120 15 -0.98 0.80 167.59
125 15 -1.48 0.71 167.64
130 15 -2.08 0.62 167.70
135 15 -2.79 0.53 167.77
140 15 -3.64 0.43 167.84
145 15 -4.65 0.34 167.91
150 15 -5.86 0.26 167.98
155 15 -7.33 0.18 168.04
160 15 -9.19 0.12 168.10
165 15 -11.62 0.07 168.15
170 15 -15.09 0.03 168.18
175 15 -21.08 0.01 168.21
180 15 -80.83 0.00 179.59
0 20 -80.83 0.00 -0.41
5 20 -21.10 0.01 168.18
10 20 -15.10 0.03 168.17
15 20 -11.62 0.07 168.14
20 20 -9.19 0.12 168.09
25 20 -7.34 0.18 168.04
30 20 -5.86 0.26 167.97
35 20 -4.65 0.34 167.91
40 20 -3.64 0.43 167.83
45 20 -2.79 0.53 167.76
50 20 -2.08 0.62 167.70
55 20 -1.48 0.71 167.64
60 20 -0.98 0.80 167.59
65 20 -0.57 0.88 167.55
70 20 -0.24 0.95 167.53
75 20 0.01 1.00 167.51
80 20 0.18 1.04 167.50
85 20 0.29 1.07 167.50
90 20 0.32 1.08 167.50
95 20 0.29 1.07 167.50
100 20 0.18 1.04 167.50
105 20 0.01 1.00 167.51
110 20 -0.24 0.95 167.53
115 20 -0.57 0.88 167.56
120 20 -0.98 0.80 167.59
125 20 -1.48 0.71 167.64
130 20 -2.08 0.62 167.70
135 20 -2.79 0.53 167.77
140 20 -3.64 0.43 167.84
145 20 -4.65 0.34 167.91
150 20 -5.86 0.26 167.98
155 20 -7.33 0.18 168.04
160 20 -9.19 0.12 168.10
165 20 -11.62 0.07 168.15
170 20 -15.09 0.03 168.18
175 20 -21.08 0.01 168.21
180 20 -80.83 0.00 179.59
0 25 -80.83 0.00 -0.41
5 25 -21.10 0.01 168.18
10 25 -15.10 0.03 168.17
15 25 -11.62 0.07 168.14
20 25 -9.19 0.12 168.09
25 25 -7.34 0.18 168.04
30 25 -5.86 0.26 167.97
35 25 -4.65 0.34 167.91
40 25 -3.64 0.43 167.83
45 25 -2.79 0.53 167.76
50 25 -2.08 0.62 167.70
55 25 -1.48 0.71 167.64
60 25 -0.98 0.80 167.59
65 25 -0.57 0.88 167.55
70 25 -0.24 0.95 167.53
75 25 0.01 1.00 167.51
80 25 0.18 1.04 167.50
85 25 0.29 1.07 167.50
90 25 0.32 1.08 167.50
95 25 0.29 1.07 167.50
100 25 0.18 1.04 167.50
105 25 0.01 1.00 167.51
110 25 -0.24 0.95 167.53
115 25 -0.57 0.88 167.56
120 25 -0.98 0.80 167.59
125 25 -1.48 0.71 167.64
130 25 -2.08 0.62 167.70
135 25 -2.79 0.53 167.77
140 25 -3.64 0.43 167.84
145 25 -4.65 0.34 167.91
150 25 -5.86 0.26 167.98
155 25 -7.33 0.18 168.04
160 25 -9.19 0.12 168.10
165 25 -11.62 0.07 168.15
170 25 -15.09 0.03 168.18
175 25 -21.08 0.01 168.21
180 25 -80.83 0.00 179.59
0 30 -80.83 0.00 -0.41
5 30 -21.10 0.01 168.19
10 30 -15.10 0.03 168.17
15 30 -11.62 0.07 168.14
20 30 -9.19 0.12 168.09
25 30 -7.34 0.18 168.04
30 30 -5.86 0.26 167.98
35 30 -4.65 0.34 167.91
40 30 -3.64 0.43 167.83
45 30 -2.79 0.53 167.76
50 30 -2.08 0.62 167.70
55 30 -1.48 0.71 167.64
60 30 -0.98 0.80 167.59
65 30 -0.57 0.88 167.55
70 30 -0.24 0.95 167.53
75 30 0.01 1.00 167.51
80 30 0.18 1.04 167.50
85 30 0.29 1.07 167.50
90 30 0.32 1.08 167.50
95 30 0.29 1.07 167.50
100 30 0.18 1.04 167.51
105 30 0.01 1.00 167.51
110 30 -0.24 0.95 167.53
115 30 -0.57 0.88 167.56
120 30 -0.98 0.80 167.59
125 30 -1.48 0.71 167.64
130 30 -2.08 0.62 167.70
135 30 -2.79 0.53 167.77
140 30 -3.64 0.43 167.84
145 30 -4.65 0.34 167.91
150 30 -5.86 0.26 167.98
155 30 -7.33 0.18 168.04
160 30 -9.19 0.12 168.10
165 30 -11.62 0.07 168.15
170 30 -15.09 0.03 168.18
175 30 -21.08 0.01 168.21
180 30 -80.83 0.00 179.59
0 35 -80.83 0.00 -0.41
5 35 -21.10 0.01 168.19
10 35 -15.10 0.03 168.17
15 35 -11.62 0.07 168.14
20 35 -9.19 0.12 168.09
25 35 -7.34 0.18 168.04
30 35 -5.86 0.26 167.98
35 35 -4.65 0.34 167.91
40 35 -3.64 0.43 167.83
45 35 -2.79 0.53 167.76
50 35 -2.08 0.62 167.70
55 35 -1.48 0.71 167.64
60 35 -0.98 0.80 167.59
65 35 -0.57 0.88 167.56
70 35 -0.24 0.95 167.53
75 35 0.01 1.00 167.51
80 35 0.18 1.04 167.50
85 35 0.29 1.07 167.50
90 35 0.32 1.08 167.50
95 35 0.29 1.07 167.50
100 35 0.18 1.04 167.51
105 35 0.01 1.00 167.51
110 35 -0.24 0.95 167.53
115 35 -0.57 0.88 167.56
120 35 -0.98 0.80 167.59
125 35 -1.48 0.71 167.64
130 35 -2.08 0.62 167.70
135 35 -2.79 0.53 167.77
140 35 -3.64 0.43 167.84
145 35 -4.65 0.34 167.91
150 35 -5.86 0.26 167.98
155 35 -7.33 0.18 168.04
160 35 -9.18 0.12 168.10
165 35 -11.62 0.07 168.15
170 35 -15.09 0.03 168.18
175 35 -21.08 0.01 168.21
180 35 -80.83 0.00 179.59
0 40 -80.83 0.00 -0.41
... 159721 more lines ...
|
| Attachment 2: setup.txt
|
///////////////////
NFOUR=1024
EXPONENT=21
NNU=100 // number of neutrino events
NNU_PASSED=10 // number of neutrino events that are allowed to pass the trigger
ONLY_PASSED_EVENTS=1 // 0 (default): AraSim throws NNU events whether or not they pass; 1: AraSim throws events until the number of events that pass the trigger is equal to NNU_PASSED (WARNING: may cause long run times if reasonable values are not chosen)
NOISE_WAVEFORM_GENERATE_MODE=0 // generate new noise waveforms for each events
NOISE_EVENTS=16 // number of pure noise waveforms
TRIG_ANALYSIS_MODE=0 // 0 = signal + noise, 1 = signal only, 2 = noise only
DETECTOR=1 // ARA stations 1 to 7
NOFZ=1
core_x=10000
core_y=10000
TIMESTEP=5.E-10 // value for 2GHz actual station value
TRIG_WINDOW=1.E-7 // 100ns which is actual testbed trig window
POWERTHRESHOLD=-6.06 // 100Hz global trig rate for 3 out of 16 ARA stations
//DATA_BIN_SIZE=8192
POSNU_RADIUS=3000
V_MIMIC_MODE=0 // 0 : global trig is located center of readout windows
DATA_SAVE_MODE=0 // 2 : don't save any waveform informations at all
DATA_LIKE_OUTPUT=0 // 0 : don't save any waveform information to eventTree
BORE_HOLE_ANTENNA_LAYOUT=0
SECONDARIES=0
// below settings are available for only TestBed mode (DETECTOR=3 case)
TRIG_ONLY_BH_ON=0
CALPULSER_ON=0
USE_MANUAL_GAINOFFSET=0
USE_TESTBED_RFCM_ON=0
NOISE_TEMP_MODE=0
TRIG_THRES_MODE=0
READGEOM=0 // reads geometry information from the sqlite file or not (0 : don't read)
// new variables
//INTERACTION_MODE=0 // pickunbiased mode!
//taumodes=1
//BH_ANT_SEP_DIST_ON=0 // default 0 : use constant borehole antenna distance. 1 : use separate antenna distance. use z_btw01, z_btw12, ... in ARA_N_info.txt or ARA37_info.txt
TRIG_MODE=0 // use vpol, hpol separated trigger mode. by default N_TRIG_V=3, N_TRIG_H=3. You can change this values
//N_TRIG_V=4
//N_TRIG_H=4
number_of_stations=1
core_x=10000
core_y=10000
///////////////////
//DATA_SAVE_MODE=2
// Settings for Arbitrary event generation
//EVENT_TYPE=10 // arbitrary event type
//INTERACTION_MODE=2 // pick exact location using settings below
//POSNU_THETA=1.7 // interaction location elevation angle in radians
//POSNU_THETA=-0.5 // interaction location elevation angle in radians
//POSNU_PHI=0 // interaction location azimuthal angle in radians
//POSNU_R=1000 // interaction location radius in meters
//INTERACTION_MODE=4 // pick interaction location within cylindrical volume above the ice
//PICK_ABOVE_HEIGHT=3000.
// Settings for Station A2-like simulation
//DETECTOR=4
//DETECTOR_STATION=2
//DATA_LIKE_OUTPUT=1
///////////////////////// Other Alternate Settings //////////////////
//POSNU_RADIUS=5000
//RAYSOL_RANGE=7000
//SELECT_FLAVOR=1
//NU_NUBAR_SELECT_MODE=1
//SELECT_CURRENT=1 //default 2: get from ratios in Ghandi etal paper, 0: nc, 1: cc
//NNU_THIS_THETA=1
//NNU_THETA=0.785
//NNU_D_THETA=0.0873
//NNU_D_THETA=0.0
//NOISE_EVENTS=16
//NOISE_EVENTS=1
//NOISE=0
//NOISE_TEMP_MODE=1
//ANTENNA_MODE=1
//APPLY_NOISE_FIGURE=1
//ALL_ANT_V_ON=0
|
|
|
197
|
Mon Feb 20 12:33:20 2023 |
Dennis H. Calderon | AraSim Simulations for Top 5 Antennas (12/2022) with Higher Stats | AraSim Simulations for Top 5 Antennas from 12/2022 Run with 3 million simulated events
Simulated using Arasim version currently used in GENETIS Loop
/fs/ess/PAS1960/BiconeEvolutionOSC/AraSim/
AraSim ROOT files stored in
/fs/ess/PAS1960/ROOT_Files_Higher_Stats/AraSim_Loop_122022/Feb_2023_Crazy_Sides/
Antennas simulated
- Generation: 13, Individual: 84
- Generation: 18, Individual: 89
- Generation: 19, Individual: 96
- Generation: 29, Individual: 87
- Generation: 9, Individual: 50
Results
| Antenna |
Total Events |
Triggered Events |
Weighted Events |
Effective Volume |
Effective Volume Error |
| 13_84 |
2995982 |
40251 |
16058 |
5.71 |
0.035 |
| 18_89 |
3010000 |
40224 |
15963 |
5.65 |
0.035 |
| 19_96 |
3010000 |
39989 |
16034 |
5.68 |
0.031 |
| 29_87 |
3010000 |
40056 |
16049 |
5.68 |
0.034 |
| 9_50 |
3010000 |
40042 |
16057 |
5.69 |
0.034 |
| ARA_Sym_Bicone_6in |
3010000 |
36627 |
14334 |
5.08 |
0.032 |
Conclusion
Antenna from Generation 13 Individual 84 performed the best with these stats |
| Attachment 1: Crazy_Sides_Triggered_Event_Data.pdf
|
|
|
240
|
Tue May 21 09:55:57 2024 |
Jacob Weiler | AraSim CSE Spring 2024 Work | # AraSim CSE Spring 2024 Work
## Goals
The main goal was to get a working multithreaded version of the AraSim codebase working. Doing this, the hope was to learn how to multithread the code and get it in a good place to hopefully also integrate GPU's at a later date.
## Where is it at currently?
A bulk of the work done was to functionalize the Connect_Interaction_Detector_V2 to allow for multithreading and to cleanup codebase.
### Going through code added/changed
Helper functions for multiple parts of code. They went through and split it up into multiple parts. Putting line numbers when needed
Part 1: Clearning Antenna Data
Part 2: Determine gain channel
Part 3: solve ray tracing (Not Done)
Part 4: Process Ray Tracing Solution (Not Done)
Part 5: Calculate Signal Factors (Not Done)
Part 6: Calculate Antenna Gain Factors (Not Done)
Part 7: Process Frequency Domain Signal
Part 8A: Process Neutrino Events. Lines 908 - 1208 (Not Done)
Part 8B: Process Arbitrary Events. Lines 1209 - 1475
Part 8C: Process Simple Pulser Simulation. Lines 1478 - 1747 (Not Done)
Part 8D: Process PVA Pulser Simulation. Lines 1751 - 2110 (Not Done)
Part 8E: Process Calpulser Event. Lines 2113 - 2445
Part 9A: Process Noise. Lines 2593 - 2955
Part 9B: Process Trigger and Mimic Waveforms. Lines 2994 - 3624
## What still needs to be done?
- Multithreading still isn't working, multiple threads are writing data to the same place causing the program to crash. This need to be resolved to at least have a working prototype.
- I believe for multithreading we need to mark explicitly where file/data writing is happening to be able to adjust to make thread-safe
- Double checking that new functions are passing variables in the correct way. The CSE students had this has a slight fear.
- Some helper functions are still not completed (3-6, 8A, 8C, 8D)
- Current completed parts of code are all in separate branches and need to be merged after double checking that variable passing is correct |
|
|
148
|
Fri Mar 25 17:14:01 2022 |
Alex M | Antenna Minimum Length Investigation | The reviewer of the paper we are trying to publish (as well as other external colleagues we showed the paper to) asked about our minimum length we implemented in the loop. Currently, we cut off the length at a minimum of 37.5 cm for each cone (minimum of 75 cm for the full cone). There is an ELOG post from Amy that presented the reasoning for this ( http://radiorm.physics.ohio-state.edu/elog/GENETIS/81 ). We initially thought that we needed a minimum because the antennas were evolving to be small (around and lower than the minimum). We thought that XFdtd was being inaccurate at lengths lower than 37.5 cm, so we set that as a minimum. To see if we can replicate any strange behavior from XFdtd, I simulated antennas at and below our minimum length and generated plots of their antennas responses. The genes are listed below, corresponding to the patterns in the plot. The results do not look unusual (that is, not dissimilar to the other bicones we have generated, including the ARA bicone) and seem to show an improved sensitivity in the upward direction for shorter bicones. The equation we used to arrive at this minimum (f = c/(4L)) might be indicative of a maximum length rather than a minimum.
| Inner radius |
Length |
Quadratic |
Linear |
2.4892,37.5,-0.00142604,0.032832
4.64522,37.5,0.00153863,-0.14004
2.4892,34.5,-0.00142604,0.032832
4.64522,34.5,0.00153863,-0.14004
2.4892,31.5,-0.00142604,0.032832
4.64522,31.5,0.00153863,-0.14004
2.4892,27.5,-0.00142604,0.032832
4.64522,27.5,0.00153863,-0.14004
2.4892,23.5,-0.00142604,0.032832
4.64522,23.5,0.00153863,-0.14004
2.4892,19.5,-0.00142604,0.032832
4.64522,19.5,0.00153863,-0.14004
2.4892,15.5,-0.00142604,0.032832
4.64522,15.5,0.00153863,-0.14004
|
| Attachment 1: polar_plot_283.37.png
| 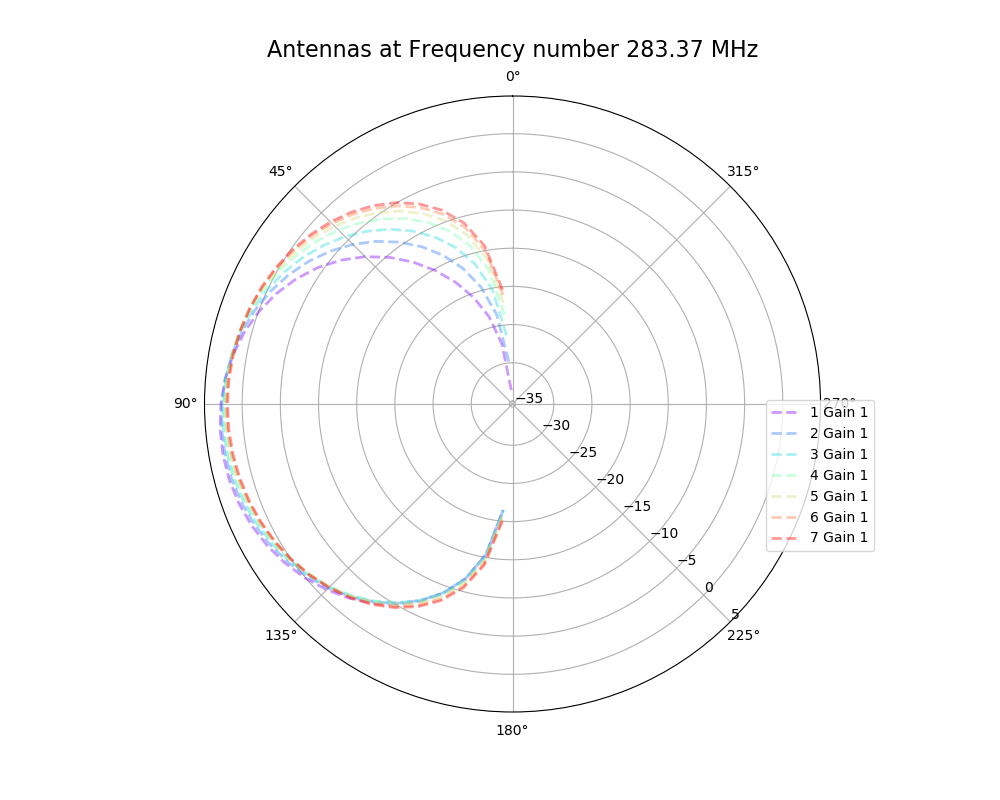 |
|
|
27
|
Thu Dec 5 14:32:54 2019 |
Julie Rolla | Adding Constraints To-Do List | In my last post, I mentioned (1) our to-do list for adding the ARA hole constraints, and (2) that we've made a mistake upon implementing it. I've attached detailed steps of that to-do list that I've written, as well as an explanation for the correction that is currently being worked on. Please see the pdf attached for an explanation of corrections.
|
| Attachment 1: AddingConstraints.pdf
|
|
|
211
|
Tue Apr 11 00:36:49 2023 |
Bryan Reynolds | AREA physics of results plot first look | After some work, I've gotten the physics of results plotting script provided by Dennis working to compare the ARASim outputs from an AREA run to a set of reference data for the ARA bicone (provided by Alex). Among the outputs is a histogram of of events triggered vs cos(theta_nu) (attached here). This is the plot that was previously presented in GENETIS talks in the physics of results discussion.
Here I compared the best individual from the AREA results (Blue- "source_2") that I have on hand (only the ARASim output files from the most recent generation ran are currently saved) with the ARA bicone reference data (Red- "source_1"). Both datasets included a total of 150,000 NNU. Note that the AREA run used was a single frequency test, meaning that the radiation pattern was held constant for all frequencies in the band.
Also attached for reference are the gain pattern of the associated individual from the AREA output and the violin plot of the run producing the data. |
| Attachment 1: 20230410_AREA_PoR_CosTheta_bestIndiv_singleFreqTest.PNG
| 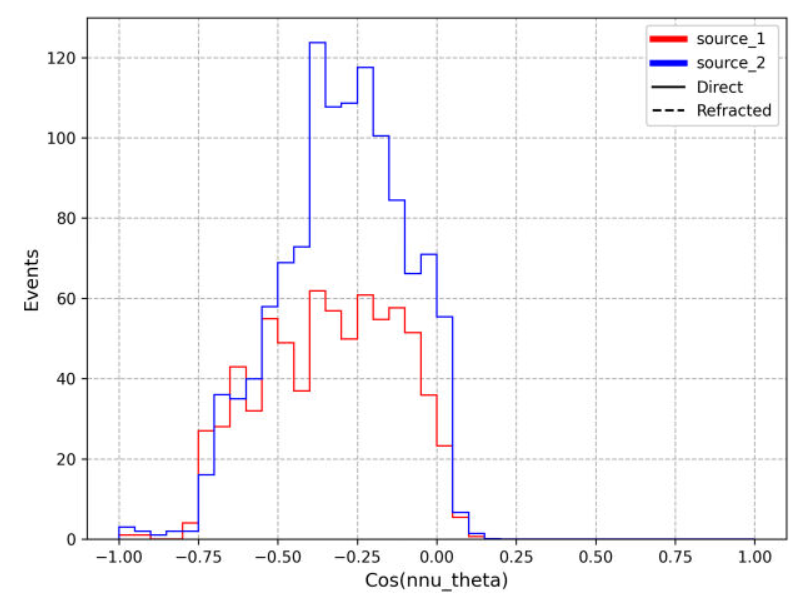 |
| Attachment 2: 20230201_singleFreqTest_100NPop_fitnessPlot_10Gens.PNG
| 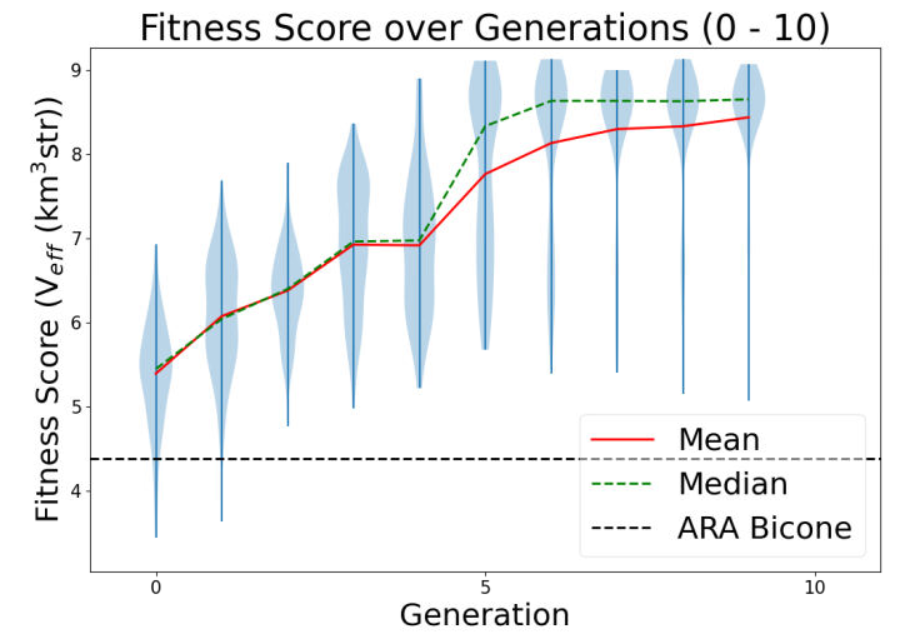 |
| Attachment 3: 20230410_AREA_mostFitIndivReUpload_singleFreqTest.PNG
| 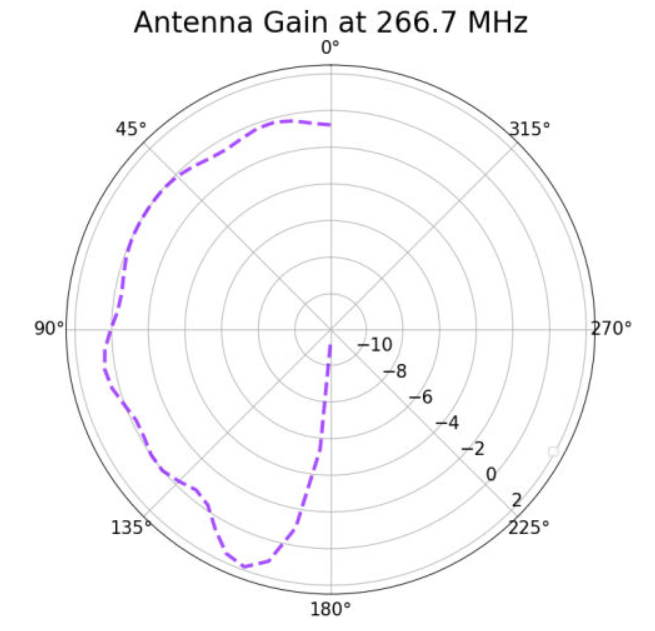 |
|
|
218
|
Thu May 11 15:57:08 2023 |
Ryan Debolt, Byran Reynolds | AREA Updates | Here is some backlogged information as well as recent updates to our progress on AREA and its optimizations:
4/13/2023
We concluded our initial test of the AREA optimization loop. While analyzing our results, we noticed that most of our runs never reached our benchmarks for performance (chi-squared scores of .25 and .1) and that those that did contain abnormally high amounts of mutation. From previous runs using the GA for the GENETIS loop, we find that mutation and immigration (Note: AREA does not use immigration) usually play a smaller role in overall growth and are simply there to promote diversity. Crossover on the other hand is what handles the bulk of the growth done by the GA. Given our results ran counter to that, we decided to look further into the matter.
Upon looking at the fitness score files, we could see that mutation-created individuals did not perform well in the best run types, but the crossover ones did contain the bulk of good scores. This lines up with expectations. However, closer inspection revealed that these crossover-derived individuals had extremely similar fitness scores, which could indicate elitism in selection methods leading to a lack of diversity in solutions. We then looked at a run that used only crossover with one selection type (roulette), and found every individual to have identical (and poor) scores that were the same up to around the 4th decimal place. This is a strong case that the selection methods used are too elitist to grow a population properly.
We dove into the AREA GA to take a look at the functions doing the selection methods, and discovered several issues potentially causing excess elitism in the GA. It seems that the roulette selection does not behave the way we have used in our other GAs. There appears to be no weighting of individuals; rather, a threshold fitness score is collected and only individuals with a fitness score above the threshold are selected. This causes elitism, as only the most fit individuals are likely to pass this threshold. Tournament was found to be similarly troubled, with a bracket size of ⅓ of the population. This is again very elitist, as it is very large, making it very likely that only the best individuals will be selected. Before we try to further optimize the AREA GA, these selection methods should be addressed and fixed.
For roulette, we propose implementing a similar weighting scheme for individuals as is used in the PAEA GAs that gives preference to the most fit individuals, but still allows others to be selected to encourage diversity of solutions.
For crossover, we propose decreasing the bracket size. This would allow a more diverse range of individuals to be selected through “winning” smaller brackets that would have a lower probability of including top performing individuals.
4/20/23
We (Bryan and Ryan) met to work on improving the selection method functions in the AREA algorithm.
For roulette selection, we added weighting by fitness score, as is done in the other GENETIS GAs, in an effort to prevent it from disproportionately selecting the most fit individuals only to become parents. As a first test, we ran an abbreviated version of the test-loop code, printing the fitness scores of the parents selected by the roulette method. The fix appears to have passed this initial test, as the fitness scores showed more variety and in the case of roulette crossover two unique parents seemed to be selected.
For tournament selection, the only change necessary appeared to be decreasing the bracket size of the tournament(s), which in turn encourages other individuals besides the most fit to be selected as parents, therefore increasing diversity of solutions. We ran out of time to test this change fully, so we will pick up here in our next meeting.
Looking further ahead, we will plan to work on refactoring the AREA algorithm to use the same functions as the other GENETIS GAs wherever possible. In the meantime, this working version can still be used for feature development so that progress is not halted.
5/11/2023
Upon looking at the results of the recent optimizations, our best runs are still taking over 40 generations to reach an optimized score. Given the speed of AraSim, this is still too slow to be considered optimal. Comparing this test loop run to the ones run on by the broader GENETIS GA, our best runs for the AREA GA are worse than the worst runs from those optimizations. As such, we have decided to pause optimizing the AREA GA in favor of adding it to the broader GENETIS one. As of today, we have finished a rough version of the generating function necessary to create new individuals. We have also started progress on the constraining functions and scaling functions that will be necessary to generate the spherical harmonics used to find the gain and phase. Once these functions are complete, we should be able to transplant this GA back into our test loop for AREA and resume optimizations.
|
|
|
151
|
Fri Apr 8 16:06:31 2022 |
Aidan Snyder | AREA - frequency linear dependence correlation test run - 04/05/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- to see if the linear dependence on frequency was properly implemented
- What is different about this run from the last?
- Under closer examination, the previous run of AREA appeared to not linearly depend on frequency, so we recompiled the GA in order to fix this
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Number of neutrinos
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
- Result
- There is a problem with how the GA has assigned the Veff values, resulting in many individuals being assigned zero Veff, however the linear dependence seems to have been implemented successfully
Note: We also ran a run called 20220405fahimi5run1, which was a short test run with a very low amount of individuals and neutrinos which seemed to work fine; as in the linear dependence seemed to work. |
|
|
146
|
Fri Feb 11 16:34:54 2022 |
Aidan Snyder | AREA - frequency linear dependence correlation test run - 01/18/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- to see if the linear dependence on frequency was properly implemented
- What is different about this run from the last?
- attempted to evolve gain pattern as a linear function of frequency
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
|
| Attachment 1: Screenshot_2022-02-11_164514.png
| 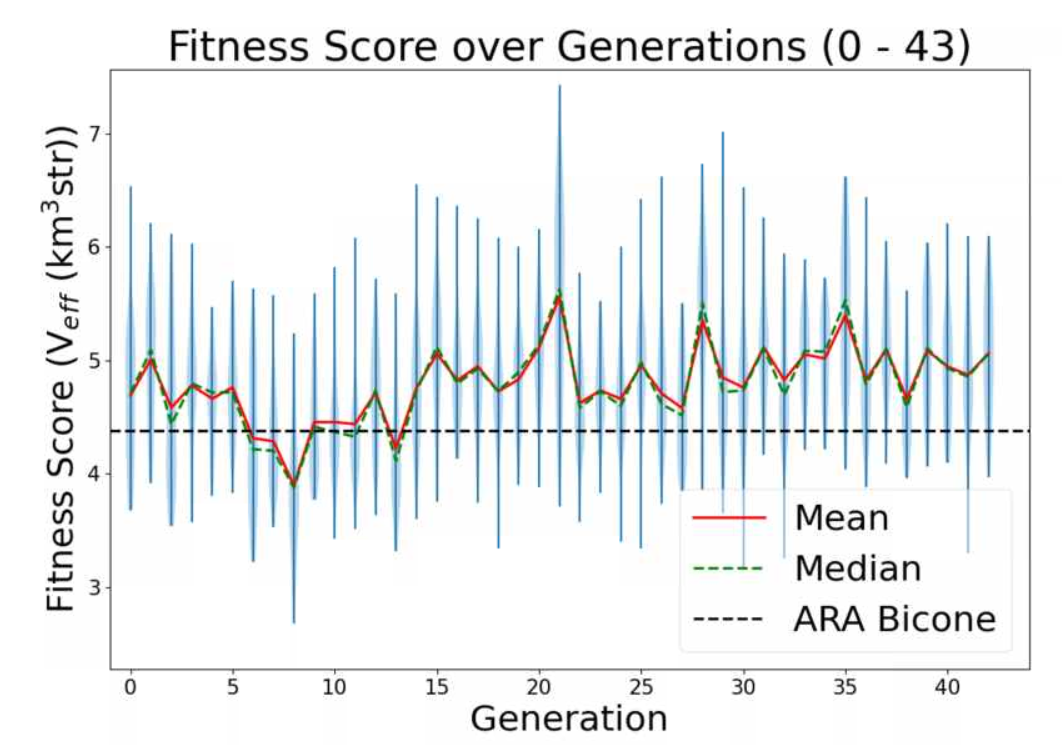 |
| Attachment 2: Screenshot_2022-02-11_164136.png
| 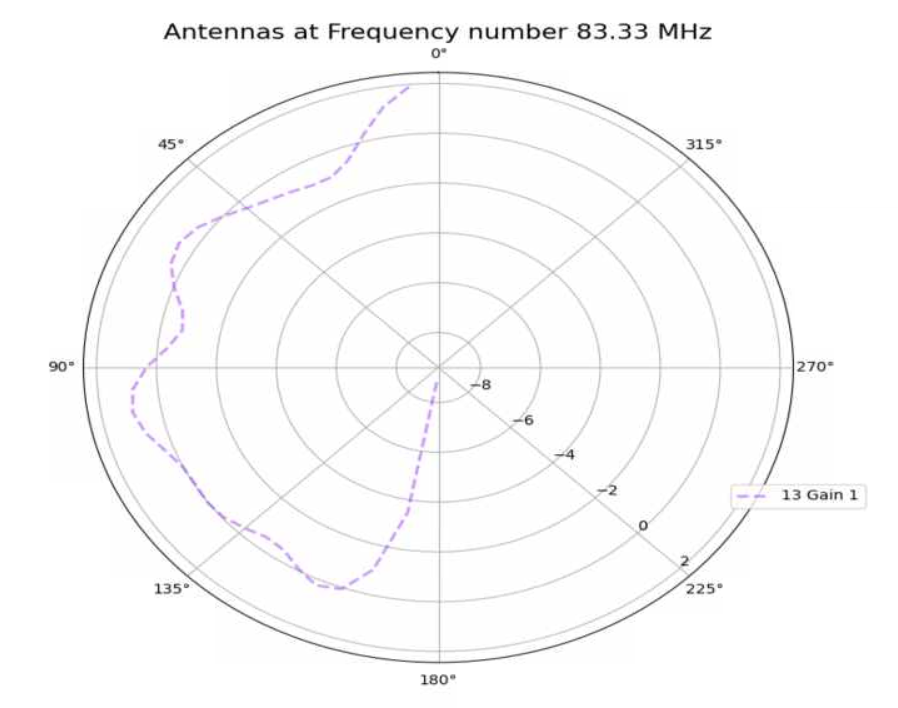 |
| Attachment 3: Screenshot_2022-02-11_164252.png
| 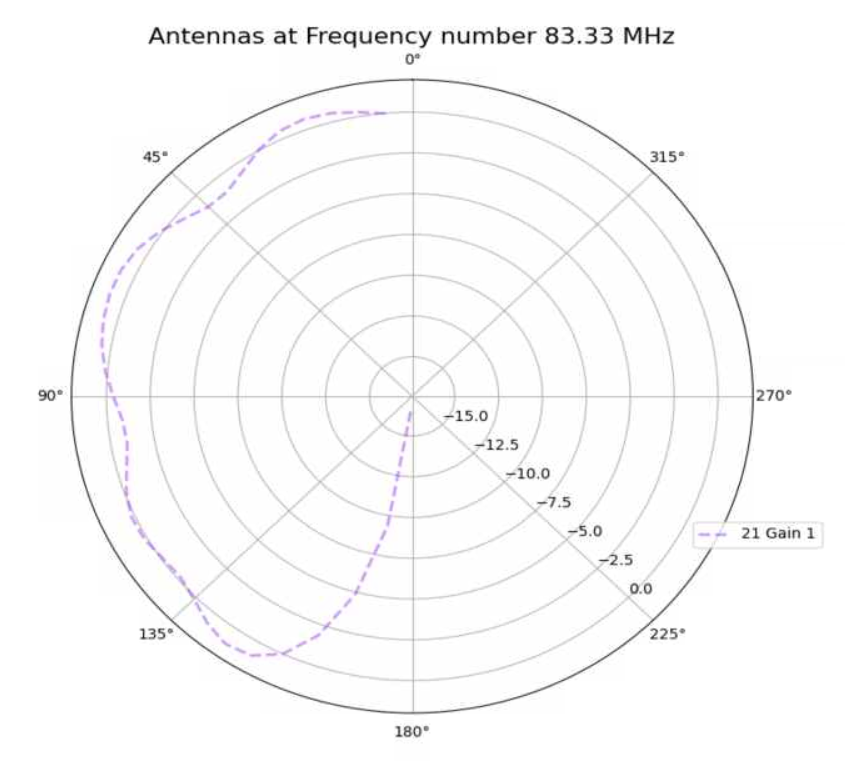 |
| Attachment 4: Screenshot_2022-02-11_164406.png
| 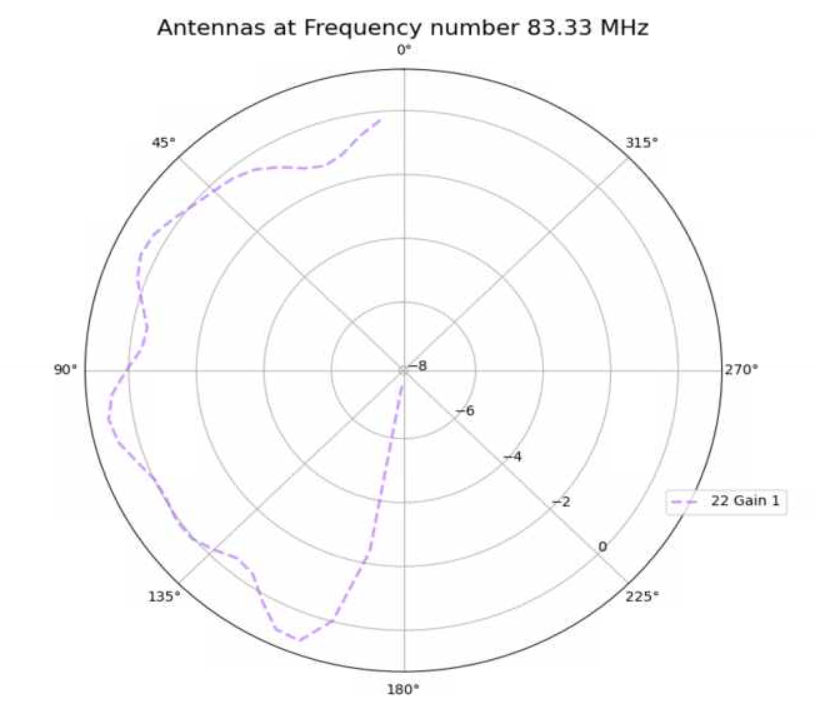 |
|
|
153
|
Tue Apr 19 17:46:07 2022 |
Aidan Snyder | AREA - Short run testing stringReplacement2.py - 04/19/2022 |
- Run Type
- Run Date
- Run Name
- Why are we doing this run?
- Check to see if stringReplacement2.py changes were successfull
- What is different about this run from the last?
- added below code in order to create a fitness file inside of each generation to display the fitness scores for just that generation, as opposed to just having one big file full of all the generations.
f2 = open(source + "/" + "gen_{}".format(gen) + "/fitnessFile_gen_{}".format(gen) + ".txt", 'a')
f2.write(','.join(mean_Veff_array) + "\n")
f2.close()
- Symmetric, asymmetric, linear, nonlinear?
- Number of individuals (NPOP)
- Operators / Selection methods used (% of each)
- roulette crossover 50%
- roulette mutation 16%
- tournament crossover 18%
- tournament mutation 16%
- Are we using the database?
|
|
|
190
|
Tue Jan 31 15:31:29 2023 |
Alex M | ARA Bicone Responses | Here we will record where we obtained the ARA bicone antenna response files we use as our baselines. We want to record where we found them/who gave them to us and how they were generated (either through simulation or from actual tests). |
|
|
15
|
Tue Jul 2 13:16:30 2019 |
Julie Rolla | APS slides | The following are links to the 2018 and 2018 APS slides.
2018 APS talk:
https://www.dropbox.com/home/GP_Antennas/APS%20Talk?preview=April+APS+Talk_final.pdf
2019 APS talk:
https://absuploads.aps.org/presentation.cfm?pid=15183 |
|
|
202
|
Mon Mar 20 12:37:38 2023 |
Amy | ANITA/PUEO pictures | Here are some ANITA/PUEO pictures that I received from Christian Mike that might be helpful. I include his descriptions as well.
The one with the black crates in the background is the ANITA 1 and ANITA 2 form factor and the one in the anechoic chamber is the ANITA 3 and ANITA 4 form factor. I dimensioned out an ANITA 3/4 antenna, in CAD space, to give you a better idea of the ridge profile. The ANITA 1/2 CAD models we have, I found, are inaccurate.
Attached are images of the PUEO antenna array geometry. One image annotates the approximate locations of the Vpol feed with the top antenna feed as the origin.
|
| Attachment 1: IMG_2362.jpg
|  |
| Attachment 2: IMG_2364.jpg
|  |
| Attachment 3: IMG_2365.jpg
|  |
|
|
81
|
Thu Jul 2 23:13:05 2020 |
Amy | A useful reference on bicone antennas | I found this datasheet on bicone antennas, that is a useful reference for seeing the relationship between bicone dimensions and bandwidth. From these it looks like the lower end of the band is at about f_low=c/(4*L_E), where L_E is the entire length of the antenna (twice the length of one cone), although we'd expect the peak gain to be where a quarter wavelength is just one of the cones (and the figure may roughly support that).
The attached file comes from this website:
https://www.ramayes.com/Biconical_Antennas.htm
|
| Attachment 1: biconall.pdf
|
|
|
10
|
Tue Mar 5 16:36:51 2019 |
Julie Rolla | 3/5 Task Updates | Tue Mar 5 17:29:02 2019
The things we worked on this week is in bold. Below the bold items is a description of exactly what we took care of.
- Paper:
- Julie added Amy's edits.
- Julie let Kai know he is welcome to edit, too.
- Julie will also keep edits updated as they come in!
- Get loop working-- specifically XF:
- Cade has now officially gotten reasonable outputs for his XF scripts. See the attached images for the XF outputs and associated CAD drawing. It seems to look like a reasonable gain pattern.
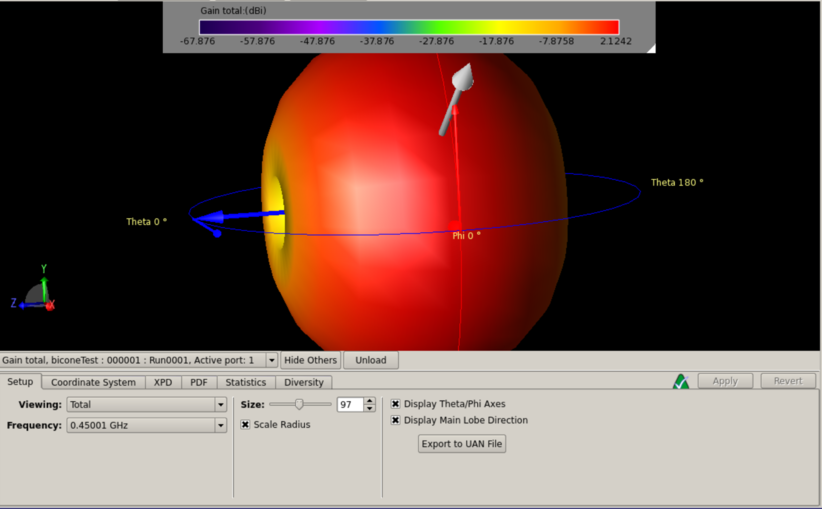
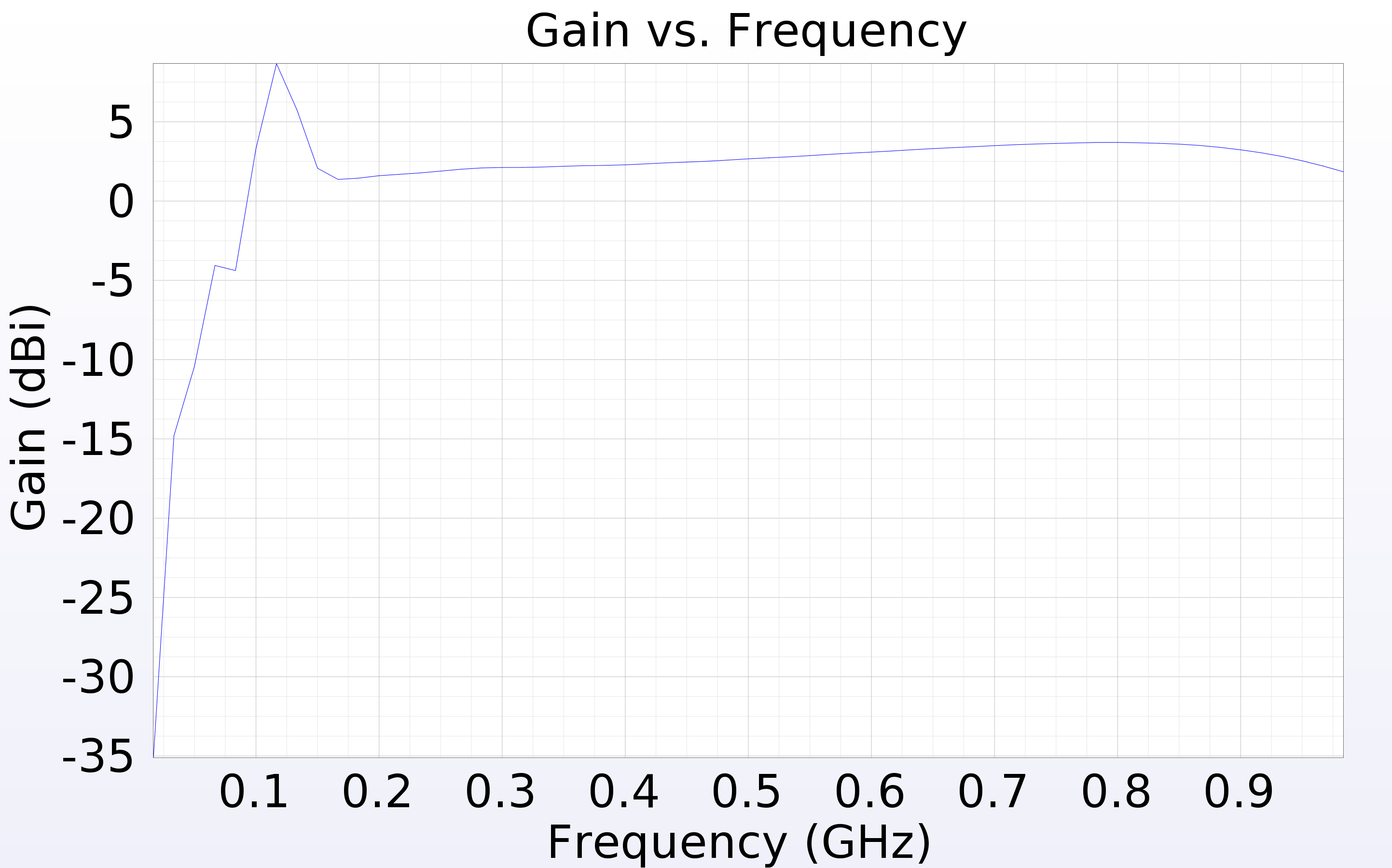

- The loop now works until AraSim. We are now getting the following AraSim errors:
- XF_Loop.sh: line 244: ./AraSim: No such file or directory
- XF_Loop.sh: line 244: ./AraSim: No such file or directory
- Julie is going to help Cade make sure that this directory is correct and that it has been compiled! More info to come.
- Test GA (Suren):
- I've been working on creating a gaussian fitness score with which I can test the performance of our genetic algorithm. The idea is as follows: We run the GA and get a generation of individuals. We provide their fitness score by simply taking the values {radius, length, angle} as the {x, y, z} coordinate in a 3D space. We can then use a 3D gaussian to output a value at that location, and use that number as our fitness score. The beauty of this method is that we can then plot the progress of our genetic algorithm each generation as a dot in this 3D space, and we can see it slowly snake to the center. Additionally, we can use more complex fitness scores, like one with a local maxima and a global maxima to see if it get's stuck in the local maxima. Below, I have an image I generated on python of a 2D section contour plot of a 3D gaussian in length-angle space. The dark purple is a lower value and the yellow is a higher value of the gaussian. The lines and dots hand-drawn on top are an idea of what it would look for like to start the GA at the red dot and see it's progress each generation until it reaches the center, shown by the white dots. Running this for our current roulette algorithm would make a very nice GA proof of concept plot for the paper, I believe!
.png.png)
- Gain pattern plots -- Make them 3D (Evelyn) & put on GitHub
- We need to figure out how to make a group Github. We have a lot of Githubs made by people and we need them all accessible from a group account. Right now the old dipole Github and the CalPoly stuff are linked to a group account. We need to find out how to get the Bicone account held by Max, and Evelyn's plotting code all on the group account: https://github.com/gp-antennas.
- Update: Evelyn and Max have contacted Brian to get their accounts linked. It looks like Amy, Brian, and Stephanie (Cal Poly) are the owners.
- Insert LR plot & Gain plot to loop (?)
- Edit bash script
- Update Github
- AraSim plots (Julie and Max) -- see #2 and #3 here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/7 for details.
- Max and Julie have gotten sample data on Owens.
- located at /users/PCON0003/cond0068/ARA/AraSim/branches/AraSimLite/outputs
- Max and Julie have gotten sample programs from Jorge.
- https://github.com/toej93/AraSim_noise_calib/blob/master/readTree.cc
- https://github.com/toej93/AraSim_noise_calib/blob/master/M.readTree
- Julie installed root, and had Max source Julie's thisroot.sh files to run root on Owens.
- Max installed Arasim to his user.
- Next step: run plotting software given by Jorge.
- Update manual with things from Suren's last post here: http://radiorm.physics.ohio-state.edu/elog/GENETIS/8
|
| Attachment 4: data.csv
|
Frequency (GHz),Theta (°),Phi (°),Gain phi,Gain theta
0.08333,0,0,4.19491965e-34,9.30908802e-36
0.08333,0,15.0000004,1.13745804e-34,2.83160959e-36
0.08333,0,30.0000008,2.90124587e-34,1.98981462e-34
0.08333,0,45.0000013,4.33400635e-34,3.34216749e-35
0.08333,0,60.0000017,2.87577966e-34,3.75231251e-34
0.08333,0,74.9999987,9.63247596e-35,1.39309362e-34
0.08333,0,90.0000025,2.77504471e-37,2.0774204e-34
0.08333,0,105,8.43720541e-35,1.15045184e-34
0.08333,0,120.000003,2.91249572e-34,3.04555617e-34
0.08333,0,135,2.34227902e-34,4.30943346e-34
0.08333,0,149.999997,1.06040209e-34,4.11289678e-34
0.08333,0,164.999994,7.50737618e-35,5.81275966e-36
0.08333,0,180.000005,1.78965268e-33,7.3492858e-37
0.08333,0,195.000002,2.02610741e-33,2.48287308e-35
0.08333,0,209.999999,8.89165876e-34,2.33583341e-35
0.08333,0,224.999996,3.16031794e-34,5.36348869e-34
0.08333,0,240.000007,1.47586622e-33,3.91377831e-35
0.08333,0,255.000004,2.87337932e-36,6.29063644e-34
0.08333,0,270.000001,1.31261981e-36,6.31632558e-34
0.08333,0,284.999998,1.8150178e-34,1.62607603e-34
0.08333,0,299.999995,5.04654556e-34,1.25443146e-33
0.08333,0,314.999992,1.93889207e-34,1.35787597e-34
0.08333,0,329.999989,2.98205296e-35,4.5730018e-34
0.08333,0,345.000013,2.06764199e-34,3.06302518e-35
0.08333,0,360.00001,4.19491965e-34,9.30908802e-36
0.08333,15.0000004,0,2.21307282e-12,0.0883042589
0.08333,15.0000004,15.0000004,1.24958681e-12,0.0883042738
0.08333,15.0000004,30.0000008,4.24763008e-13,0.0883042812
0.08333,15.0000004,45.0000013,1.43870359e-14,0.0883042961
0.08333,15.0000004,60.0000017,1.76734427e-13,0.0883042812
0.08333,15.0000004,74.9999987,8.30293787e-13,0.0883042738
0.08333,15.0000004,90.0000025,1.74207111e-12,0.0883042589
0.08333,15.0000004,105,2.68639538e-12,0.0883042738
0.08333,15.0000004,120.000003,3.48234115e-12,0.0883042812
0.08333,15.0000004,135,3.94334608e-12,0.0883042961
0.08333,15.0000004,149.999997,3.88229336e-12,0.0883042812
0.08333,15.0000004,164.999994,3.2406417e-12,0.0883042738
0.08333,15.0000004,180.000005,2.21092025e-12,0.0883042589
0.08333,15.0000004,195.000002,1.15119351e-12,0.0883042738
0.08333,15.0000004,209.999999,3.67093942e-13,0.0883042812
0.08333,15.0000004,224.999996,1.35828941e-14,0.0883042961
0.08333,15.0000004,240.000007,1.44884037e-13,0.0883042812
0.08333,15.0000004,255.000004,7.57448195e-13,0.0883042738
0.08333,15.0000004,270.000001,1.743383e-12,0.0883042589
0.08333,15.0000004,284.999998,2.82562493e-12,0.0883042738
0.08333,15.0000004,299.999995,3.63690545e-12,0.0883042812
0.08333,15.0000004,314.999992,3.93473708e-12,0.0883042961
0.08333,15.0000004,329.999989,3.70870695e-12,0.0883042812
0.08333,15.0000004,345.000013,3.08593168e-12,0.0883042738
0.08333,15.0000004,360.00001,2.21307282e-12,0.0883042589
0.08333,30.0000008,0,3.04286353e-11,0.330554247
0.08333,30.0000008,15.0000004,1.77038245e-11,0.330554426
0.08333,30.0000008,30.0000008,6.15558559e-12,0.330554664
0.08333,30.0000008,45.0000013,1.95957738e-13,0.330554813
0.08333,30.0000008,60.0000017,2.64734307e-12,0.330554664
0.08333,30.0000008,74.9999987,1.186425e-11,0.330554426
0.08333,30.0000008,90.0000025,2.39625351e-11,0.330554247
0.08333,30.0000008,105,3.62047961e-11,0.330554426
0.08333,30.0000008,120.000003,4.70035712e-11,0.330554664
0.08333,30.0000008,135,5.41321606e-11,0.330554813
0.08333,30.0000008,149.999997,5.42172245e-11,0.330554664
0.08333,30.0000008,164.999994,4.53593517e-11,0.330554426
0.08333,30.0000008,180.000005,3.04133004e-11,0.330554247
0.08333,30.0000008,195.000002,1.54130996e-11,0.330554426
0.08333,30.0000008,209.999999,4.83360053e-12,0.330554664
0.08333,30.0000008,224.999996,1.90661912e-13,0.330554813
0.08333,30.0000008,240.000007,1.83763486e-12,0.330554664
0.08333,30.0000008,255.000004,1.00491724e-11,0.330554426
0.08333,30.0000008,270.000001,2.39718714e-11,0.330554247
0.08333,30.0000008,284.999998,3.96109014e-11,0.330554426
0.08333,30.0000008,299.999995,5.08507958e-11,0.330554664
0.08333,30.0000008,314.999992,5.40794388e-11,0.330554813
0.08333,30.0000008,329.999989,5.01597201e-11,0.330554664
0.08333,30.0000008,345.000013,4.16781852e-11,0.330554426
0.08333,30.0000008,360.00001,3.04286353e-11,0.330554247
0.08333,45.0000013,0,1.19610113e-10,0.663833082
0.08333,45.0000013,15.0000004,7.04909117e-11,0.663833678
0.08333,45.0000013,30.0000008,2.47605096e-11,0.663835049
0.08333,45.0000013,45.0000013,7.7185464e-13,0.663835585
0.08333,45.0000013,60.0000017,1.07718721e-11,0.663835049
0.08333,45.0000013,74.9999987,4.7389391e-11,0.663833797
0.08333,45.0000013,90.0000025,9.42051992e-11,0.663833082
0.08333,45.0000013,105,1.41075526e-10,0.663833797
0.08333,45.0000013,120.000003,1.83183593e-10,0.663835049
0.08333,45.0000013,135,2.12397419e-10,0.663835585
0.08333,45.0000013,149.999997,2.1427396e-10,0.663835049
0.08333,45.0000013,164.999994,1.79498763e-10,0.663833678
0.08333,45.0000013,180.000005,1.19567467e-10,0.663833082
0.08333,45.0000013,195.000002,5.99928648e-11,0.663833678
0.08333,45.0000013,209.999999,1.8719401e-11,0.663835049
0.08333,45.0000013,224.999996,7.59097917e-13,0.663835585
0.08333,45.0000013,240.000007,6.99973326e-12,0.663835049
0.08333,45.0000013,255.000004,3.89681343e-11,0.663833797
0.08333,45.0000013,270.000001,9.42311576e-11,0.663833082
0.08333,45.0000013,284.999998,1.56817601e-10,0.663833797
0.08333,45.0000013,299.999995,2.01015038e-10,0.663835049
0.08333,45.0000013,314.999992,2.1228902e-10,0.663835585
0.08333,45.0000013,329.999989,1.95689701e-10,0.663835049
0.08333,45.0000013,345.000013,1.62577174e-10,0.663833678
0.08333,45.0000013,360.00001,1.19610113e-10,0.663833082
0.08333,60.0000017,0,2.64464534e-10,0.999831378
0.08333,60.0000017,15.0000004,1.54434562e-10,0.999833047
0.08333,60.0000017,30.0000008,5.38681218e-11,0.999836028
0.08333,60.0000017,45.0000013,1.71404438e-12,0.999837577
0.08333,60.0000017,60.0000017,2.32321419e-11,0.999836028
0.08333,60.0000017,74.9999987,1.03637522e-10,0.999833047
0.08333,60.0000017,90.0000025,2.08304943e-10,0.999831676
0.08333,60.0000017,105,3.1357697e-10,0.999833047
0.08333,60.0000017,120.000003,4.06610884e-10,0.999836028
0.08333,60.0000017,135,4.68799e-10,0.999837577
0.08333,60.0000017,149.999997,4.70491313e-10,0.999836028
0.08333,60.0000017,164.999994,3.9417325e-10,0.999833047
0.08333,60.0000017,180.000005,2.64387595e-10,0.999831378
0.08333,60.0000017,195.000002,1.34046177e-10,0.999833047
0.08333,60.0000017,209.999999,4.21460956e-11,0.999836028
0.08333,60.0000017,224.999996,1.69582761e-12,0.999837577
0.08333,60.0000017,240.000007,1.5866277e-11,0.999836028
0.08333,60.0000017,255.000004,8.72237838e-11,0.999833047
0.08333,60.0000017,270.000001,2.08351794e-10,0.999831676
0.08333,60.0000017,284.999998,3.44218515e-10,0.999833047
0.08333,60.0000017,299.999995,4.41347736e-10,0.999836028
0.08333,60.0000017,314.999992,4.68693473e-10,0.999837577
0.08333,60.0000017,329.999989,4.3446402e-10,0.999836028
0.08333,60.0000017,345.000013,3.6132225e-10,0.999833047
0.08333,60.0000017,360.00001,2.64464534e-10,0.999831378
0.08333,74.9999987,0,4.04069306e-10,1.24752045
0.08333,74.9999987,15.0000004,2.29066002e-10,1.24752307
0.08333,74.9999987,30.0000008,7.80540424e-11,1.24752772
0.08333,74.9999987,45.0000013,2.62711142e-12,1.2475301
0.08333,74.9999987,60.0000017,3.28389468e-11,1.24752796
0.08333,74.9999987,74.9999987,1.52820298e-10,1.24752307
0.08333,74.9999987,90.0000025,3.18272853e-10,1.24752045
0.08333,74.9999987,105,4.88081409e-10,1.24752307
0.08333,74.9999987,120.000003,6.3120531e-10,1.24752796
0.08333,74.9999987,135,7.15345061e-10,1.2475301
0.08333,74.9999987,149.999997,7.06045777e-10,1.24752772
0.08333,74.9999987,164.999994,5.90874849e-10,1.24752307
0.08333,74.9999987,180.000005,4.03963918e-10,1.24752045
0.08333,74.9999987,195.000002,2.1085797e-10,1.24752307
0.08333,74.9999987,209.999999,6.75901626e-11,1.24752772
0.08333,74.9999987,224.999996,2.61011547e-12,1.2475301
0.08333,74.9999987,240.000007,2.62591095e-11,1.24752796
0.08333,74.9999987,255.000004,1.3817425e-10,1.24752307
0.08333,74.9999987,270.000001,3.18337051e-10,1.24752045
0.08333,74.9999987,284.999998,5.15459897e-10,1.24752307
0.08333,74.9999987,299.999995,6.62257138e-10,1.24752796
0.08333,74.9999987,314.999992,7.15343729e-10,1.2475301
0.08333,74.9999987,329.999989,6.74007739e-10,1.24752772
0.08333,74.9999987,345.000013,5.61640512e-10,1.24752307
0.08333,74.9999987,360.00001,4.04069306e-10,1.24752045
0.08333,90.0000025,0,4.61999022e-10,1.33854401
0.08333,90.0000025,15.0000004,2.51170362e-10,1.33854699
0.08333,90.0000025,30.0000008,8.27910379e-11,1.33855271
0.08333,90.0000025,45.0000013,3.00353275e-12,1.33855546
0.08333,90.0000025,60.0000017,3.37403577e-11,1.33855271
0.08333,90.0000025,74.9999987,1.66223049e-10,1.33854699
0.08333,90.0000025,90.0000025,3.639051e-10,1.33854425
0.08333,90.0000025,105,5.73373904e-10,1.33854699
0.08333,90.0000025,120.000003,7.39050543e-10,1.33855271
0.08333,90.0000025,135,8.17466539e-10,1.33855546
0.08333,90.0000025,149.999997,7.88037857e-10,1.33855271
0.08333,90.0000025,164.999994,6.58218591e-10,1.33854699
0.08333,90.0000025,180.000005,4.61882532e-10,1.33854401
0.08333,90.0000025,195.000002,2.51093729e-10,1.33854699
0.08333,90.0000025,209.999999,8.27514446e-11,1.33855271
0.08333,90.0000025,224.999996,2.99441093e-12,1.33855546
0.08333,90.0000025,240.000007,3.37565774e-11,1.33855271
0.08333,90.0000025,255.000004,1.66264363e-10,1.33854699
0.08333,90.0000025,270.000001,3.63976016e-10,1.33854425
0.08333,90.0000025,284.999998,5.73479764e-10,1.33854699
0.08333,90.0000025,299.999995,7.3919082e-10,1.33855271
0.08333,90.0000025,314.999992,8.17630574e-10,1.33855546
0.08333,90.0000025,329.999989,7.88206e-10,1.33855271
0.08333,90.0000025,345.000013,6.58368415e-10,1.33854699
0.08333,90.0000025,360.00001,4.61999022e-10,1.33854401
0.08333,105,0,4.04069306e-10,1.24752057
0.08333,105,15.0000004,2.10708131e-10,1.24752307
0.08333,105,30.0000008,6.72303047e-11,1.24752772
0.08333,105,45.0000013,2.61616965e-12,1.2475301
0.08333,105,60.0000017,2.66377094e-11,1.24752796
0.08333,105,74.9999987,1.38355521e-10,1.24752307
0.08333,105,90.0000025,3.1827288e-10,1.24752045
0.08333,105,105,5.15625764e-10,1.24752307
0.08333,105,120.000003,6.62567279e-10,1.24752796
0.08333,105,135,7.15196014e-10,1.2475301
0.08333,105,149.999997,6.73421985e-10,1.24752772
0.08333,105,164.999994,5.61246716e-10,1.24752307
0.08333,105,180.000005,4.03963918e-10,1.24752057
0.08333,105,195.000002,2.28775943e-10,1.24752307
0.08333,105,209.999999,7.76212219e-11,1.24752772
0.08333,105,224.999996,2.61669375e-12,1.2475301
0.08333,105,240.000007,3.32482444e-11,1.24752796
0.08333,105,255.000004,1.530778e-10,1.24752307
0.08333,105,270.000001,3.18337051e-10,1.24752045
0.08333,105,284.999998,4.88436347e-10,1.24752307
0.08333,105,299.999995,6.31764474e-10,1.24752796
0.08333,105,314.999992,7.15488113e-10,1.2475301
0.08333,105,329.999989,7.0575884e-10,1.24752772
0.08333,105,345.000013,5.90749838e-10,1.24752307
0.08333,105,360.00001,4.04069306e-10,1.24752057
0.08333,120.000003,0,2.64464478e-10,0.999831378
0.08333,120.000003,15.0000004,1.33883321e-10,0.999832869
0.08333,120.000003,30.0000008,4.17817655e-11,0.999835908
0.08333,120.000003,45.0000013,1.69906689e-12,0.999837399
0.08333,120.000003,60.0000017,1.6242372e-11,0.999835908
0.08333,120.000003,74.9999987,8.74086845e-11,0.999832869
0.08333,120.000003,90.0000025,2.08304943e-10,0.999831378
0.08333,120.000003,105,3.444173e-10,0.999832869
0.08333,120.000003,120.000003,4.41701259e-10,0.999835908
0.08333,120.000003,135,4.68590555e-10,0.999837399
0.08333,120.000003,149.999997,4.33917985e-10,0.999835908
0.08333,120.000003,164.999994,3.60962066e-10,0.999832869
0.08333,120.000003,180.000005,2.6438754e-10,0.999831378
0.08333,120.000003,195.000002,1.5416636e-10,0.999832869
0.08333,120.000003,209.999999,5.34479926e-11,0.999835908
0.08333,120.000003,224.999996,1.70535331e-12,0.999837399
0.08333,120.000003,240.000007,2.36332828e-11,0.999835908
0.08333,120.000003,255.000004,1.03881029e-10,0.999832869
0.08333,120.000003,270.000001,2.08351794e-10,0.999831378
0.08333,120.000003,284.999998,3.13908594e-10,0.999832869
0.08333,120.000003,299.999995,4.07135714e-10,0.999835908
0.08333,120.000003,314.999992,4.68895589e-10,0.999837399
0.08333,120.000003,329.999989,4.70152972e-10,0.999835908
0.08333,120.000003,345.000013,3.94003968e-10,0.999832869
0.08333,120.000003,360.00001,2.64464478e-10,0.999831378
0.08333,135,0,1.19610072e-10,0.663833082
0.08333,135,15.0000004,5.99381239e-11,0.663833678
0.08333,135,30.0000008,1.85767825e-11,0.663835049
0.08333,135,45.0000013,7.60673751e-13,0.663835585
0.08333,135,60.0000017,7.14823036e-12,0.663835049
0.08333,135,74.9999987,3.90346644e-11,0.663833678
0.08333,135,90.0000025,9.42051992e-11,0.663833082
0.08333,135,105,1.56898619e-10,0.663833678
0.08333,135,120.000003,2.01159506e-10,0.663835049
0.08333,135,135,2.12236353e-10,0.663835585
0.08333,135,149.999997,1.95445715e-10,0.663835049
0.08333,135,164.999994,1.62410418e-10,0.663833678
0.08333,135,180.000005,1.19567467e-10,0.663833082
0.08333,135,195.000002,7.03755873e-11,0.663833678
0.08333,135,209.999999,2.45850649e-11,0.663835049
0.08333,135,224.999996,7.66894252e-13,0.663835585
0.08333,135,240.000007,1.09362398e-11,0.663835049
0.08333,135,255.000004,4.74905774e-11,0.663833678
0.08333,135,270.000001,9.42311437e-11,0.663833082
0.08333,135,284.999998,1.41226114e-10,0.663833678
0.08333,135,299.999995,1.83415158e-10,0.663835049
0.08333,135,314.999992,2.12445991e-10,0.663835585
0.08333,135,329.999989,2.14137152e-10,0.663835049
0.08333,135,345.000013,1.7943369e-10,0.663833678
0.08333,135,360.00001,1.19610072e-10,0.663833082
0.08333,149.999997,0,3.04286353e-11,0.330554277
0.08333,149.999997,15.0000004,1.54137154e-11,0.330554426
0.08333,149.999997,30.0000008,4.81777248e-12,0.330554783
0.08333,149.999997,45.0000013,1.91342805e-13,0.330554903
0.08333,149.999997,60.0000017,1.85569854e-12,0.330554783
0.08333,149.999997,74.9999987,1.00528414e-11,0.330554485
0.08333,149.999997,90.0000025,2.39625386e-11,0.330554277
0.08333,149.999997,105,3.9619117e-11,0.330554485
0.08333,149.999997,120.000003,5.08673659e-11,0.330554783
0.08333,149.999997,135,5.40622512e-11,0.330554903
0.08333,149.999997,149.999997,5.01101279e-11,0.330554783
0.08333,149.999997,164.999994,4.16404619e-11,0.330554426
0.08333,149.999997,180.000005,3.04133142e-11,0.330554277
0.08333,149.999997,195.000002,1.76818525e-11,0.330554426
0.08333,149.999997,209.999999,6.12725886e-12,0.330554783
0.08333,149.999997,224.999996,1.94313573e-13,0.330554903
0.08333,149.999997,240.000007,2.67184018e-12,0.330554783
0.08333,149.999997,255.000004,1.18811861e-11,0.330554485
0.08333,149.999997,270.000001,2.39718748e-11,0.330554277
0.08333,149.999997,284.999998,3.62366179e-11,0.330554485
0.08333,149.999997,299.999995,4.70487156e-11,0.330554783
0.08333,149.999997,314.999992,5.41480506e-11,0.330554903
0.08333,149.999997,329.999989,5.42033987e-11,0.330554783
0.08333,149.999997,345.000013,4.53567808e-11,0.330554426
0.08333,149.999997,360.00001,3.04286353e-11,0.330554277
0.08333,164.999994,0,2.21307608e-12,0.088304311
0.08333,164.999994,15.0000004,1.15286968e-12,0.0883043185
0.08333,164.999994,30.0000008,3.67804555e-13,0.0883043483
0.08333,164.999994,45.0000013,1.37255726e-14,0.0883043557
0.08333,164.999994,60.0000017,1.44566515e-13,0.0883043483
0.08333,164.999994,74.9999987,7.56416577e-13,0.0883043334
0.08333,164.999994,90.0000025,1.74207382e-12,0.0883043185
0.08333,164.999994,105,2.82473741e-12,0.0883043334
0.08333,164.999994,120.000003,3.63594441e-12,0.0883043483
0.08333,164.999994,135,3.93251621e-12,0.0883043557
0.08333,164.999994,149.999997,3.70533595e-12,0.0883043483
0.08333,164.999994,164.999994,3.08280896e-12,0.0883043185
0.08333,164.999994,180.000005,2.21092307e-12,0.088304311
0.08333,164.999994,195.000002,1.24800875e-12,0.0883043185
0.08333,164.999994,209.999999,4.23646713e-13,0.0883043483
0.08333,164.999994,224.999996,1.42058225e-14,0.0883043557
0.08333,164.999994,240.000007,1.77395032e-13,0.0883043483
0.08333,164.999994,255.000004,8.31207607e-13,0.0883043334
0.08333,164.999994,270.000001,1.74338549e-12,0.0883043185
0.08333,164.999994,284.999998,2.68868153e-12,0.0883043334
0.08333,164.999994,299.999995,3.48511237e-12,0.0883043483
0.08333,164.999994,314.999992,3.94543035e-12,0.0883043557
0.08333,164.999994,329.999989,3.88365945e-12,0.0883043483
0.08333,164.999994,345.000013,3.24231375e-12,0.0883043185
... 19202 more lines ...
|
|
|
Draft
|
Mon Nov 8 17:04:30 2021 |
Ethan Fahimi | 11/04/2021 AREA run 2 violin plot | This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation. |
| Attachment 1: 20211104fahimi5run2.png
| 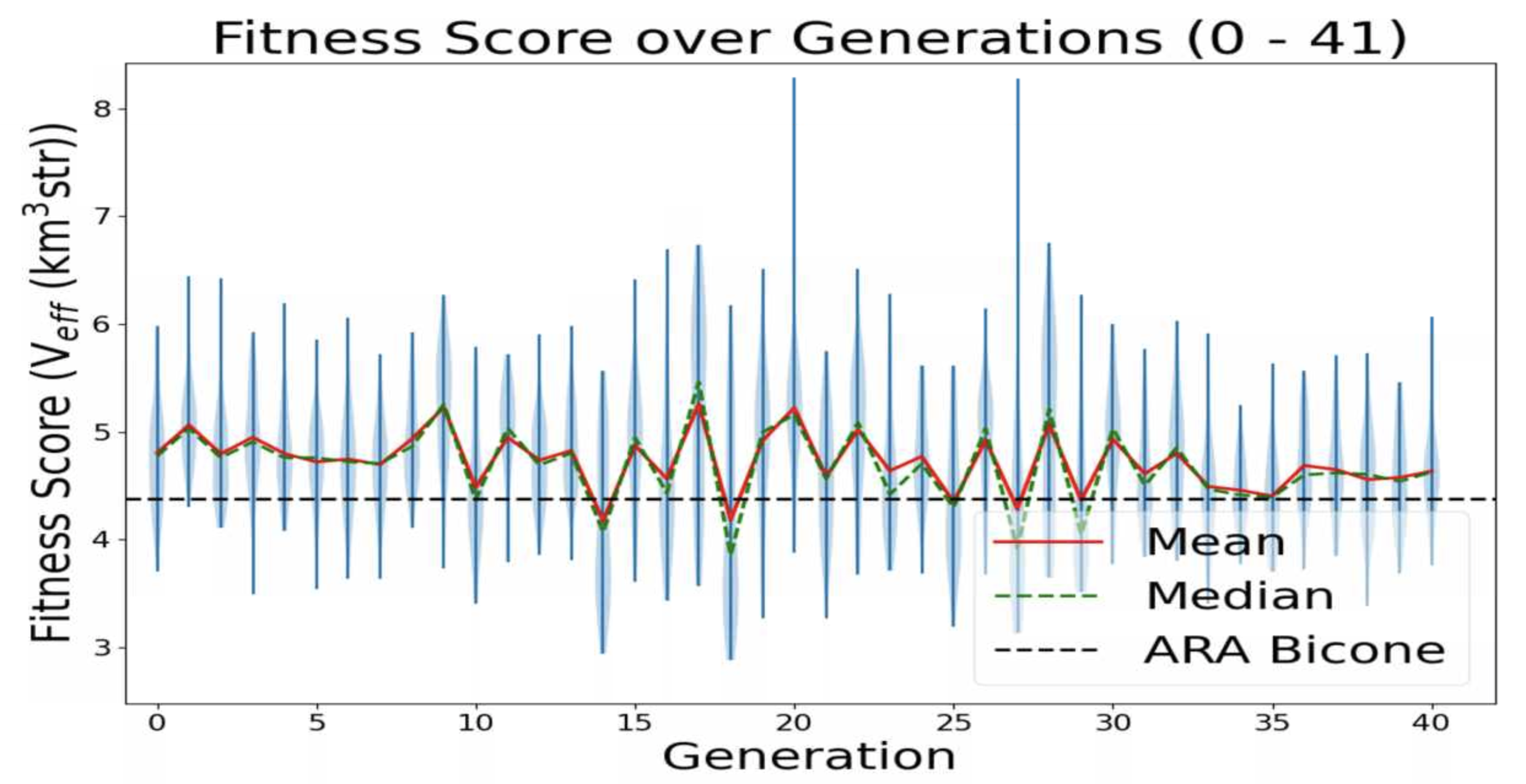 |
|
|
138
|
Fri Sep 17 13:41:36 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot | This is a plot made from the AREA project with full Arasim implementation. It can be seen that the Veff of any individuals is not what I would consider "good", nor is it really rising, it is quite flat. This is because in this version of AREA, the gain pattern at each frequency is generated differently than each other frequency, there is no correlation. This is known and actively being corrected. This plot is of old data and was just made for two reasons: to make sure that the violin plotting script works for AREA, to display this early form of AREA that has been adapted for full Arasim.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
This plot is further detailed in Julie Rolla's doctorate thesis. |
| Attachment 1: Image_9-13-21_at_6.35_PM.jpg
| 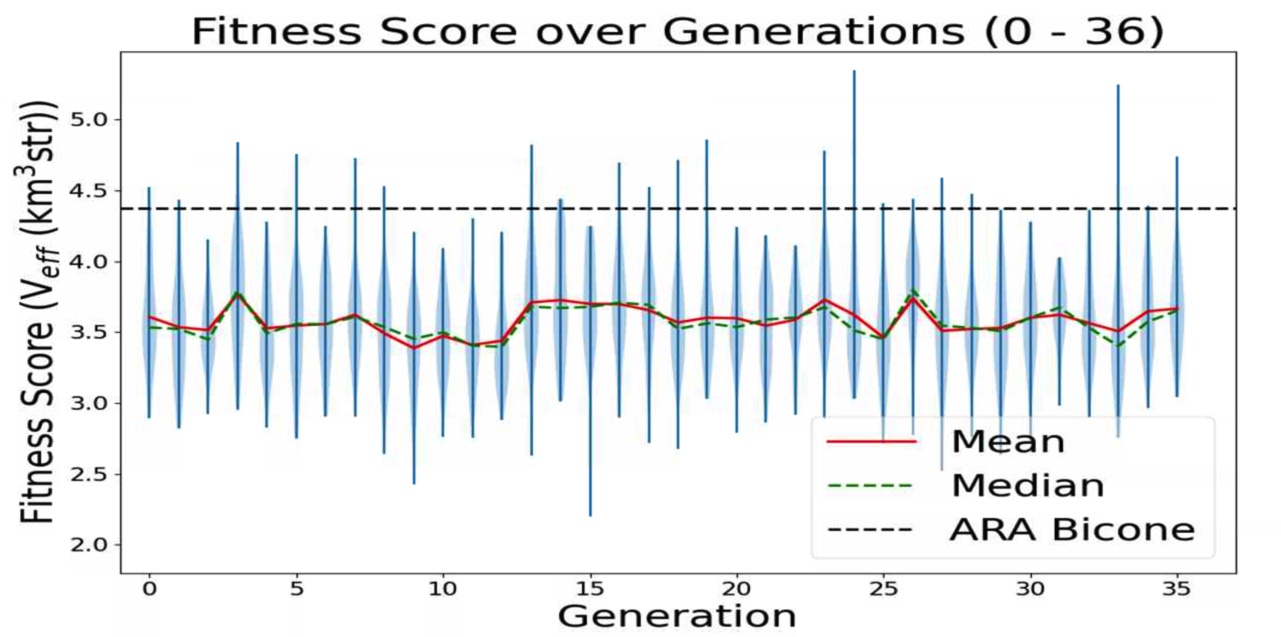 |
|
|
140
|
Mon Nov 8 17:27:01 2021 |
Ethan Fahimi | 07/20/2021 AREA run 3 violin plot | This is a plot made from the AREA project with full Arasim implementation with each gain pattern of each individual being fixed across all frequencies.
This run was done with 50 total individuals per generation, across 36 generations. Each individual was tested with 4 seeds of 10,000 neutrinos, for a total of 40,000 neutrinos. For each new generation, 25 individuals were created with roulette crossover, 8 with roulette mutation, 9 with tournament crossover, and 8 with tournament mutation.
individual 32 in gen 20 and individual 35 in gen 27 look promising (they have Veff > 8) |
| Attachment 1: 20211104fahimi5run2.png
|  |
|
|
216
|
Tue May 9 18:09:35 2023 |
Alex M | 05/08/23 Pueo Run | Here are the run details for the latest PUEO run we've started. There are still some bugs that need to be worked out, which are detailed on the github. Here is the directory where the data is stored: /fs/ess/PAS1960/HornEvolutionOSC/GENETIS_PUEO/BiconeEvolution/current_antenna_evo_build/XF_Loop/Evolutionary_Loop/Run_Outputs/2023_05_08
This run uses 50 individuals per generation with 400k neutrinos per individual at an energy exponent of 19. Only the inner side length and outer sidelength are being evolved (which evolves the height since the opening angle is fixed).
Issues With Part E (Dylan):
The Plots were not automatically created for the first generation. After a quick look through, I found that the fitness score and v effective csv files were being moved before they were copied, so the files didn’t end up in the correct place. Additionally, the errorbars.csv file was not being created, so I made a temporary fix to create a file of 0 error bars to allow for plotting the values while I work on getting the real error values.
Update: I have modified Will’s script for reading the root file outputs, so it can now read them to output a CSV file with the effective volume along with its positive and negative errors. This script does the same and more as the fitnessfunctionPUEO.py, so I will replace it in the loop when there is downtime. Additionally, this script means we don’t need to output a veff.csv file from pueoSim. I will now work on creating a less “janky” version of my pueoSim change for reading in the gain files, so I can get Will to officially add it to pueoSim and we can pull any updates in the future.
Update, Implementing the new script into Part E:
The rootAnalysis.py script is very picky with environment variables. So, first I had to load and then unload a version of python to make sure the default version is loaded. Then, I moved the sourcing of set_plotting_env.sh to later in the script to avoid having an alternate version of root sourced while running (this would cause errors). Then I source the environment for pueosim (path hardcoded, change when loop isn’t running) and loop through the rootAnalysis.py program for each individual in the generation. This will output a ${gen}_fitnessScores.csv and a ${gen}_errorBars.csv in the run directory for plotting script to use. Because it outputs these automatically, I commented out the old methods we used to create these files and removed the temporary error bar fix as well.
Another Update:
We paused the loop. I changed the hardcoded parts, and we pushed the new updates to the github. I will make comments on the changes later today.
Wednesday:
It seems like the loop is still not creating the csv files or plots for generation 1 or 2. Alex gave me the error message. The python script rootAnalysis.py had an issue with the include Geoid.h line. This was caused by sourcing the wrong things while trying to get the script working. I found that I only changed my local pueoSim environment to include the fftw installation to the $PATH and $LD_LIBRARY_PATH variables. So, I changed /fs/ess/PAS1960/buildingPueoSim/set_env.sh as well. Also, I found that the include statements in rootAnalysis.py were missing the PAS1960 portion of the path. After fixing these, and adding a missing ‘/’ to the output file path, I manually ran the rootAnalysis.py program for the 2023_05_08 run, mimicking the portion of the bash script that runs it in Part_E. It outputted the correct files in the correct format. I then ran the FScorePlot script. The median line for the ViolinPlot seemed wrong, and the FScorePlot was too scrunched with a y start of 0; I will need to change these.
Plotting Update (Dylan):
Ok, plots still aren’t being made, and I’m not running the loop, so I don’t have access to the error messages, so I’ll go through everything one by one. For Veff_Plots, this isn’t being made as there are currently no vEffectives.csv. This isn’t a big deal, as it’s just a copy of FScorePlot, but I added the creation of vEffectives.csv to rootAnalysis.py anyway. Next, I noticed that runData.csv, which is required for a couple of the plots was not being made. I looked, and the version of gensData.py had the old values for the skiprows when reading in the fitnessScores and generationDNA files, something that was overlooked when we merged the version I was developing with the version Alex was developing on. I can’t access the directory or the file, I think this is why it was overlooked in the merge. I created a temporary PUEO version in Antenna_Performance_Metric (I’m also fine with it just living here permanently).
Also, the csv files are now in the GenerationData/ directory in the run directory? This affects every plotting script and they will not work without changes. I don’t know why this was changed or if this change is permanent?
Next, ok apparently, VariablePlots isn’t even a file in the loop? We really need to use GitHub better.
I moved my local version of the program into the loop and it ran fine manually.
I am forsaking the Generation_Data directory, so I changed the rainbowPlotter and dataConverter scripts to read in a location to match all the other plotting scripts. They both also ran fine manually.
Note: The rainbowPlotter script should be able to create a gradient from the minimum fitness score to the maximum fitness score. (currently, the range is hardcoded and this can cause basically a mono-color graph.)
FScorePlot should be good
The bash script was inputting 5 variables into colorplots when the pueo version only takes in 4 (this is also something that was already fixed before the merge.) It also requires the vEffective.csv files. After copying over the fitnessScore.csv files to vEffective.csv files, it ran fine. (The vEffective.csv files will be made automatically by rootAnalysis.py in the future) |
| Attachment 1: run_details.txt
|
####### VARIABLES: LINES TO CHECK OVER WHEN STARTING A NEW RUN ###############################################################################################
RunName='2023_05_08' ## This is the name of the run. You need to make a unique name each time you run.
TotalGens=10 ## number of generations (after initial) to run through
NPOP=50 ## number of individuals per generation; please keep this value below 99
Seeds=10 ## This is how many AraSim jobs will run for each individual## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
FREQ=60 ## the number frequencies being iterated over in XF (Currectly only affects the output.xmacro loop)
NNT=40000 ## Number of Neutrinos Thrown in AraSim
exp=19 ## exponent of the energy for the neutrinos in AraSim
ScaleFactor=1.0 ## ScaleFactor used when punishing fitness scores of antennae larger than the drilling holes
GeoFactor=1 ## This is the number by which we are scaling DOWN our antennas. This is passed to many files
num_keys=6 ## how many XF keys we are letting this run use
database_flag=0 ## 0 if not using the database, 1 if using the database
## These next 3 define the symmetry of the cones.
PUEO=1 ## IF 1, we evolve the PUEO quad-ridged horn antenna, if 0, we evolve the Bicone
RADIUS=1 ## If 1, radius is asymmetric. If 0, radius is symmetric
LENGTH=1 ## If 1, length is asymmetric. If 0, length is symmetric
ANGLE=1 ## If 1, angle is asymmetric. If 0, angle is symmetric
CURVED=1 ## If 1, evolve curved sides. If 0, sides are straight
A=1 ## If 1, A is asymmetric
B=1 ## If 1, B is asymmetric
SEPARATION=0 ## If 1, separation evolves. If 0, separation is constant
NSECTIONS=2 ## The number of chromosomes
DEBUG_MODE=0 ## 1 for testing (ex: send specific seeds), 0 for real runs
## These next variables are the values passed to the GA
REPRODUCTION=3 ## Number (not fraction!) of individuals formed through reproduction
CROSSOVER=36 ## Number (not fraction!) of individuals formed through crossover
MUTATION=1 ## Probability of mutation (divided by 100)
SIGMA=5 ## Standard deviation for the mutation operation (divided by 100)
ROULETTE=20 ## Percent of individuals selected through roulette (divided by 10)
TOURNAMENT=20 ## Percent of individuals selected through tournament (divided by 10)
RANK=60 ## Percent of individuals selected through rank (divided by 10)
ELITE=0 ## Elite function on/off (1/0)
|
|
|
Draft
|
Wed May 20 10:58:38 2020 |
| | Today's Summer 2020 daily update:
As a note, today OSC was down so productivity was more limited
| Name |
Update for Today |
Plans for Tomorrow |
| Alex M. |
Mostly just wrote more on the paper in the Genetic Algorithm section. I added some citation that we used in ICRC but there are still more places that should have citations.
|
I might check tonight when OSC is back up to try to push in more updates to the loop because I wanna get Evelyn and Ryan started on running the loop. Putting in those fixes is a big priority because we want to be able to correct the potential issue with XF simulation folders being overwritten and thus uan data not corresponding to the write individuals. The two big things for me in the loop are getting the simulation data to save correctly (and also putting that in the database) and testing that we can replicate results using the specific seed. I'll probably only focus on loop stuff tomorrow.
|
| Alex P. |
Got up before OSC was down to check progress of overnight run, it seems to have worked but I noticed a problem with the database that it wasn't writing to it probably due to a permissions issue but I would have to run another time to see. Shouldn't have affected data but just the use of the database. Run got up to 8 generations with non-zero fitness score which is positive and seems to have fixed the error we originally encountered. Talked to Eliot about pointers and possible errors but was unable to look at the specific error because it is on OSC.
|
Tomorrow plan is to continue to work on database functionality and continue run to get more generations, also want to add the ability to add more plots than just the fitness score to the dropbox automatically. Plots: upload all plots (Fitness, LRT, vEff), remove legend, upload penalized red/green plot too, take off legend, add units to Fitness |
| Leo |
|
|
| Eliot |
Read about pointers and vectors in C++. Talked to Alex P a bit, and have some ideas of things to change to get the GA running. Began reading about antennas. Mostly a down day due to OSC being down.
|
Will implement changes to GA and continue familiarizing myself with how XF reads these values.
|
| Evelyn |
|
|
| Ryan |
|
|
|